Analysing a Closed Testing Procedure
AnalyseCTP.RdCalculation of p-values of a closed testing procedure (CTP).
The function returns an object of oldClass "ctp"; summary() and Display() can be applied.
AnalyseCTP(ctp.struc, model, data, factor.name = NULL, test.name = "F", ...)
Arguments
| ctp.struc | Object generated by the function |
|---|---|
| model | model of the form response~treatment. If |
| data | Dataframe, missing values in the response or treatment variable are not allowed! |
| factor.name | Character string naming the factor whose levels are compared (treatment factor). By default the first variable of the right-hand side of the model formula is used. |
| test.name | One of the following strings
|
| ... | Additional arguments for the chosen test. |
Value
An object of old class(ctp), consisting of a list with:
CTPparms: List with objects describing the CTP setup.pvalues: Dataframe with all tested hypotheses, raw and adjusted p-values.
Details
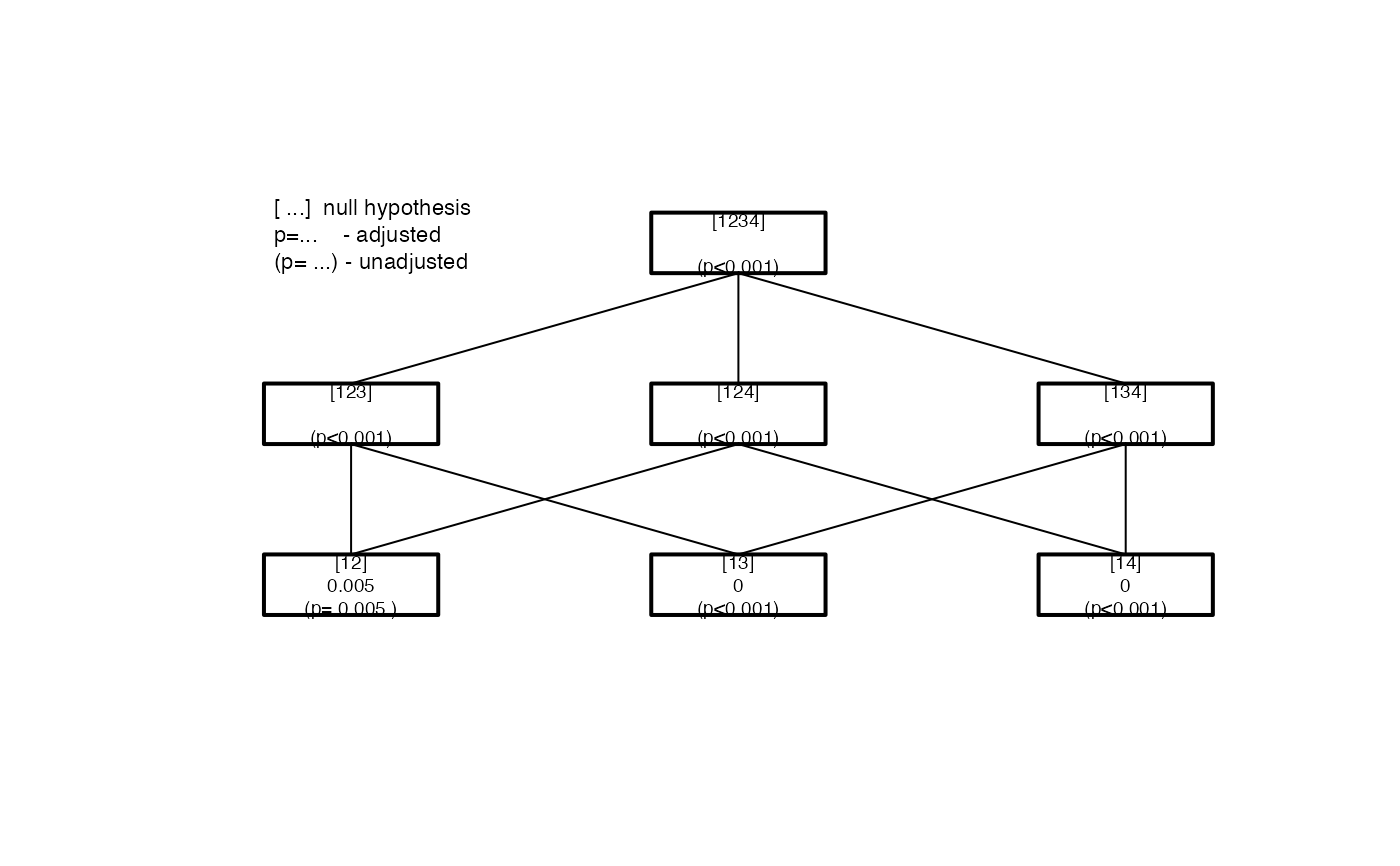
The hypothesis tree of the closed testing procedure must be created using IntersectHypotheses. For more details on the theory and the implementation as well
for many examples, see the vignettes.
Note
This procedure is constructed for testing differences and two-sided hypotheses, but not for equivalence tests. It is further based on independent samples from the population involved (i.e. on parallel group designs, but not on cross-over designs).
See also
Examples
#> #> Summary of Closed Testing Procedure #> =================================== #> #> Model : pasi.ch ~ dose , test : F #> #> Factor levels: 1=Placebo, 2=ET.10mg, 3=ET.25mg, 4=ET.50mg #> #> Elementary Hypotheses and p-values #> ---------------------------------- #> #> Hypothesis raw p-value adj. p-value #> [12] 4.748e-03 4.748e-03 #> [13] 2.737e-05 1.063e-04 #> [14] 1.826e-06 9.797e-06