2 R packages
openstatsware Workshop: Good Software Engineering Practice for R Packages
Phil
October 16, 2023
Introduction
What You Know Already
- Packages provide a mechanism for loading optional code, data, and documentation
- A library is a directory into which packages are installed
install.packages()is used to install packages into the librarylibrary()is used to load and attach packages from the library- “Attach” means that the package is put in your
searchlist — objects in the package can be used directly
- “Attach” means that the package is put in your
- Remember that package \(\neq\) library
What We Want to Talk About Now
- How to write, build, test, and check your own package 😊
- How to do this in a methodical and sustainable way
- Give tips and tricks based on practical experience
Contents of a Package
How is a Package Structured?
Package source = directory with files and subdirectories
- Mandatory:
- DESCRIPTION
- NAMESPACE
- R
- man
- Typically also includes:
- data
- inst
- src
- tests
- vignettes
- NEWS
How to Get Started Quickly
Once upon a time, developers would set up this structure manually 🥱
Nowadays, it’s super fast with:
usethis::create_package()- RStudio > File > New Project > New Directory > R Package
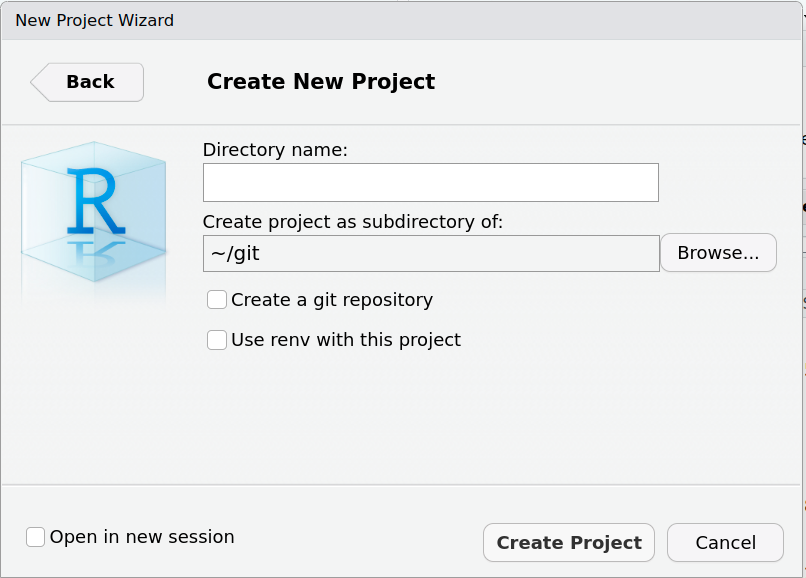
DESCRIPTION File
- Package: Choose the name of your package
- Not unimportant!
- Check CRAN to see if your name is available
- Title: Add a Title for Your Package (Title Case)
- Version: Start with a low package version
Major.Minor.Patchsyntax
- Authors@R: Add authors and maintainer
- Description: Like an abstract, including references
DESCRIPTION File (cont’d)
- License: Important for open sourcing
- Consider permissive licenses such as Apache and MIT
- Depends:
- Which R version users need to have at a minimum
- Ideally don’t put any package here
- Packages will be loaded and attached upon
libraryyour package
- Imports: Packages which you import functions, methods, classes from
- Suggests: Packages for documentation processing (
roxygen2), running examples, tests (testthat), vignettes
R Folder
- Only contains R code files (recommended to use
.Rsuffix)- Can create a file with
usethis::use_r("filename")
- Can create a file with
- Assigns R objects, i.e. mostly functions, but could also be constant variables, data sets, etc.
- Should not have any side effects, i.e. avoid
require(),options()etc. - If certain code needs to be sourced first, use on top of file (which will update the
Collatefield ofDESCRIPTIONautomatically)
NAMESPACE File
- Defines the namespace of the package, to work with R’s namespace management system
- Namespace directives in this file allow to specify:
- Which objects are exported to users and other packages
- Which are imported from other packages
NAMESPACE File (cont’d)
- Controls the search strategy for variables:
- Local (in the function body etc.)
- Package namespace
- Imports
- Base namespace
- Normal
search()path
man Folder
- Contains documentation files for the objects in the package in the
.Rdformat- The syntax is a bit similar to
LaTeX
- The syntax is a bit similar to
- All user level objects should be documented
- Internal objects don’t need to be documented — but I recommend it!
- Once upon a time, developers would set up these
.Rdfiles and theNAMESPACEmanually 🥱 - Fortunately, nowadays we have
roxygen2! 🚀
roxygen2 to the Rescue!
- We can include the documentation source directly in the R script on top of the objects we are documenting
- Syntax is composed of special comments
#'and special macros preceded with@ - In RStudio, running Build > More > Document will render the
.Rdfiles and theNAMESPACEfile for you - Get started with
usethis::use_roxygen_md() - Placing your cursor inside a function in RStudio, create a
roxygen2skeleton with Code > Insert Roxygen Skeleton
Setting up roxygen2 in your project
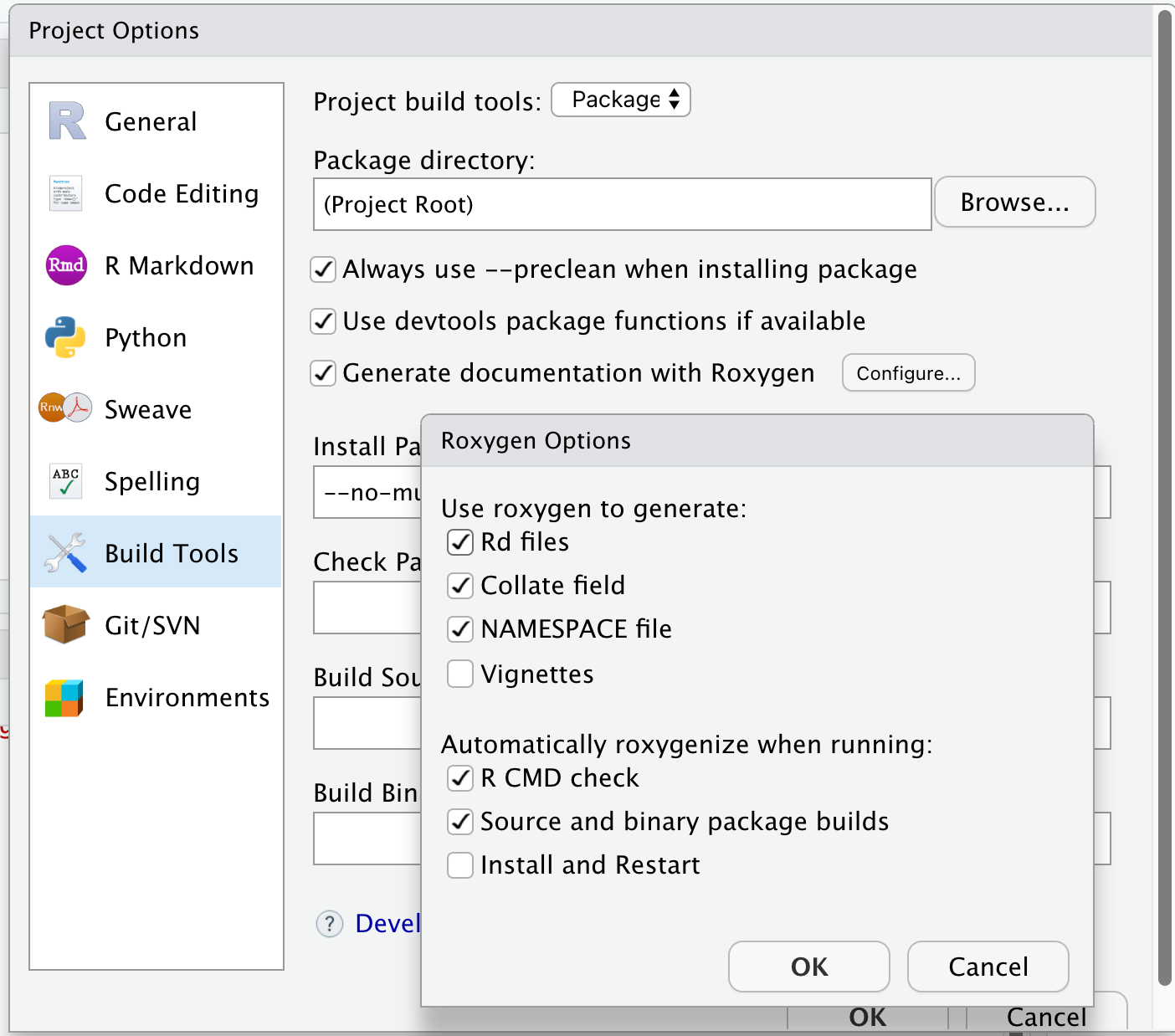
roxygen2 Source
R/my_sum.R:
#' My Summation Function
#'
#' This is my first function and it sums two numbers.
#'
#' @param x first summand.
#' @param y second summand.
#'
#' @return The sum of `x` and `y`.
#' @export
#'
#' @note This function is a bit boring but that is ok.
#' @seealso [Arithmetic] for an easier way.
#'
#' @examples
#' my_sum(1, 2)
my_sum <- function(x, y) {
x + y
}roxygen2 Output
man/my_sum.Rd:
% Generated by roxygen2: do not edit by hand
% Please edit documentation in R/bla.R
\name{my_sum}
\alias{my_sum}
\title{My Summation Function}
\usage{
my_sum(x, y)
}
\arguments{
\item{x}{first summand.}
\item{y}{second summand.}
}
\value{
The sum of \code{x} and \code{y}.
}
\description{
This is my first function and it sums two numbers.
}
\note{
This function is a bit boring but that is ok.
}
\examples{
my_sum(1, 2)
}
\seealso{
\link{Arithmetic} for an easier way.
}roxygen2 Output (cont’d)
NAMESPACE:
tests Folder
- Where store the unit tests covering the functionality of the package
- Get started with
usethis::use_testthat()andusethis::use_test()and populatetests/testthatfolder with unit tests - Rarely, tests cannot be run within
testthatframework, then these can go into R scripts directly intestsdirectory - We will look at unit tests in detail later
data Folder
- For (example) data that you ship in your package to the user
- Get started with
usethis::use_data() - Note: Usually we use lazy data loading, therefore no
data()call needed before using the data
- Get started with
- If you generate the example data, save the R script, too
- Put that into
data-rawfolder, start withusethis::use_data_raw()
- Put that into
inst Folder
- Contents will be copied recursively to installation directory
- Be careful not to interfere with standard folder names
- For data that is used by functions in the package itself
- Those would typically go into
inst/extdatafolder - Load with
system.file("path/file", package = "mypackage")
- Those would typically go into
CITATION: For customcitation()output- Create it with
usethis::use_citation()
- Create it with
inst/doccan contain documentation files (typicallypdf)
src Folder
- Contains sources and headers for any code that needs compilation
- Should only contain a single language here
- Because R uses it, mixing
C,C++andFortranusually works with OS native compilers
- Because R uses it, mixing
- Much more complex to write and maintain than an R only package
- Typically only makes sense for
- Wrapping existing libraries for use in R
- Speeding up complex computations — starting point:
Rcpp::Rcpp.package.skeleton()
vignettes Folder
- Special case of documentation files (
pdforhtml) created by compiling source files - Package users don’t need to recompile the vignettes - they are delivered with the package
- Start a new vignette with
usethis::use_vignette()- Starts an
Rmdvignette, compiled withknitr
- Starts an
- Important for the user to understand the high-level ideas
- Complements function-level documentation from our
roxygen2chunks
NEWS File
- Lists user-visible changes worth mentioning
- In each new release, add items at the top under the version they refer to
- Don’t discard old items: leave them in the file after the newer items
- Start one with
usethis::use_news_md()
Building the Package
Documenting the Package
- The first step is to produce the documentation files and
NAMESPACE - In RStudio: Build > More > Document
- In the console:
devtools::document()
Checking the Package
- R comes with pre-defined check command for packages: “the R package checker” aka
R CMD check - About 22 checks are run (so quite a lot), including things like:
- Can the package be installed?
- Is the code syntax ok?
- Is the documentation complete?
- Do tests run successfully?
- Do examples run successfully?
- In RStudio: Build > Check
- In the console:
devtools::check()
Building the Package
- The R package folder can be compressed into a single package file
- Typically we manually only build “source” package
- In RStudio: Build > More > Build Source Package
- In the console:
devtools::build()
- Makes it easy to share the package with others and submit to CRAN
Installing the Package
- R comes with pre-defined install command for packages:
R CMD INSTALL - In RStudio: Build > Install
- In the console:
devtools::install() - Note: During development it’s usually sufficient to use Build > More > Load All
- Runs
devtools::load_all() - Roughly simulates what happens when package would be installed and loaded
- Unexported objects and helpers under
testswill also be available - Key: much faster!
- Runs
Exercise
Let’s try this out now 😊
- Set up a new R package with a fancy name
- Fill out the
DESCRIPTIONfile - Include a new function
- Add
roxygen2documentation - Export the function to the namespace
- Produce the package documentation
- Run checks
- Build the package
References
License Information
- Creators (initial authors): Daniel Sabanes Bove , Liming Li
- In the current version, changes were done by (later authors): Philippe Boileau
- This work is licensed under the Creative Commons Attribution-ShareAlike 4.0 International License.
- The source files are hosted at github.com/openpharma/workshop-r-swe-mtl, which is forked from the original version at github.com/openpharma/workshop-r-swe.
- Important: To use this work you must provide the name of the creators (initial authors), a link to the material, a link to the license, and indicate if changes were made