Purpose
The staged.dependencies package simplifies the development process for developing a set of inter-dependent R packages. In each repository of the set of R packages you are co-developing you should specify a staged_dependencies.yaml file containing upstream (i.e. those packages your current repo depends on) and downstream (i.e. those packages which depend on your current repo’s package) dependency packages within your development set of packages.
The set of packages you are co-developing are internal dependencies. Other R packages they depend on are named external dependencies.
By using a git branching naming convention in your development work, staged.dependencies makes it straightforward to:
- Install upstream dependencies (and the current package): install remote upstream dependencies and the local version of the current package using the requested branches for each repository.
- Check and install downstream dependencies: install downstream and their required upstream dependencies using the requested branches for each repository (following the branch naming convention, see below). It picks up the environment variable
RCMDCHECK_ARGSand passes it as arguments toR CMD check. - Test and install downstream dependencies: see above. It tests instead of checks.
- Create dependency graphs of internal dependencies.
The package also provides:
- RStudio addins to perform the above tasks with a single click.
- Install all dependencies app: A Shiny application to explore internal dependency graphs and install packages from the graph.
Usage
Storage directory
By default, the directory ~/.staged.dependencies as well as a dummy config file are created whenever the package is loaded and the directory does not exist. This is where checked out repositories are cached. To use a different location, you can set the option staged.dependencies._storage_dir before loading the package. Note that on Windows, the path ~/.staged.dependencies may be a subdirectory of One Drive and this can lead to problems.
Note staged.dependencies requires a git signature setup that can be checked with git2r::default_signature("."). If this is not setup it can be created using git2r::config(git2r::repository("."), user.name = <<name>>, user.email = <<email>>) or via the git cli using, for example git config --global user.email <email>.
Setup tokens
If your internal packages require personal access tokens then these must be available to staged.dependencies.
There are two approaches:
- Modify your
~/.Rprofilefile with the following:
Sys.setenv("GITHUB_PAT" = "<token>") # if using https://github.com
Sys.setenv("GITLAB_PAT" = "<token>") # if using https://gitlab.com
options(
staged.dependencies.token_mapping = c(
"https://github.com" = "GITHUB_PAT",
"https://gitlab.com" = "GITLAB_PAT",
<<another_remote>> = <<token env variable>>
...
)
)- Alternatively, add your token in
~/.Renvironfile,
and add the following into your ~/.Rprofile file:
RStudio Addins
The easiest way to use staged.dependencies, once installed, is to checkout a repository and within the RStudio use the addins: 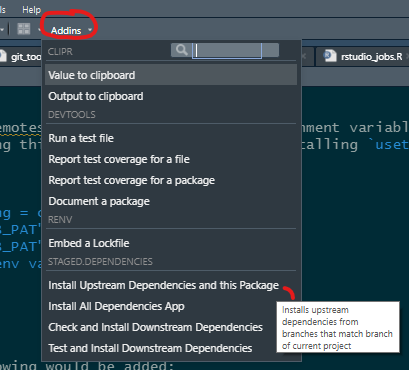
Console
You can run the actions of the addins explicitly and change the default arguments (see function documentation for further details)
# create dependency_structure object from a checked out repo...
x <- dependency_table(project = "../stageddeps.electricity", verbose = 1)
# ... or directly from a remote
x <- dependency_table(project = "openpharma/stageddeps.electricity@https://github.com",
project_type = "repo@host",
ref = "main",
verbose = 1)
# print and plot it
print(x)
if (require("visNetwork")) plot(x)
#install upstream dependencies
install_deps(x, verbose = 1)
# check downstream packages
check_downstream(x, verbose = 1, check_args = c("--no-manual"))Remember to restart the R session after installing packages that were already loaded. There are additional functions (e.g. a shiny app to view the dependency graph and choose which packages to install) in the package. See the function documentation for more details.
Additional information
Branch naming convention
staged.dependencies knows which branches to checkout for each of your projects due to a git branch convention. The default branch naming convention staged.dependencies expects throughout your repositories is described by the examples below:
- Given a branch name
a@b@c@d@main,staged.dependencieschecks for branches in each repository in the following order:a@b@c@d@main,b@c@d@main,c@d@main,d@mainandmain. - Suppose one implements a new feature on a branch called
feature1that involvesrepoB, repoCwith the dependency graphrepoA --> repoB --> repoC, whererepoAis an additional upstream dependency. One then notices thatfeature1requires a fix inrepoB, so one creates a new branchfix1@feature1@mainonrepoB. The setup can be summarized as follows:
repoA: devel
repoB: fix1@feature1@main, feature1@main, main
repoC: feature1@main, mainA PR on repoB fix1@feature1@main --> feature1@main takes repoA: main, repoB: fix1@feature1@main, repoC: feature1@main. This can be checked by setting ref = fix1@feature1@main and running check_downstream on either repoC or check_downstream on repoB (which adds repoC to its downstream dependencies). A PR on either repoB or repoC feature1@main --> main takes repoA: main, repoB:feature1@main, repoC: feature1@main, setting ref = feature1@main.
By setting ref = <<tag_name>>, staged.dependencies will checkout each repository at the tagged commit (or by default main if tag does not exist, though this can be overridden with the fallback argument to dependency_table). The check for tag name takes priority over the branch procedure described above.
Working with local packages
If you are working locally on a collection of packages, you can modify the ~/.staged.dependencies/config.yaml to automatically pick up the local packages that should be taken instead of their remote ones. The file ~/.staged.dependencies/config.yaml describes the available options. For the current project, the local rather than remote version is always taken.
Structure of staged_dependencies.yaml file
Given a feature and a git repository, the package determines the branch to checkout according to the branch naming convention described above. To get the upstream or downstream dependencies, it inspects the staged_dependencies.yaml file. This file is of the form:
---
upstream_repos:
- repo: Roche/rtables
host: https://github.com
- repo: Roche/respectables
host: https://github.com
downstream_repos:
- repo: example1/repo1
host: https://github.example.com
- repo: example1/repo2
host: https://code.example.com
current_repo:
repo: openpharma/staged.dependencies
host: https://github.comIt contains all the information that is required to fetch the repos. The current_repo lists the info to fetch the project itself. All three top level fields are required although upstream_repos and downstream_repos can be empty.
Each package is cached in a directory whose name is based on repo and a hash of repo, host. If it exists, it fetches. Otherwise, it clones. The repositories only have remote branches, the default tracking branch is deleted. Otherwise, there would be problems if the default branch is deleted from the remote, checked out locally, so git fetch --prune would not remove it and the local branch would be among the available branches considered in determine_branch.
If a repo is part of the local packages, it is not fetched, but instead copied to the cache dir. A commit is created with all files (including staged, unstaged and untracked, but excluding git-ignored) files. Given such a cache directory, the commit hash is added to the DESCRIPTION file and the package is installed, unless the package is already installed with the same commit hash. When calling install_deps and similar functions, the current project is always added to the local_repos so that its local rather than remote version is installed. For this, the current_repo field must be correct, so downstream dependencies do not fetch the remote version.
The staged_dependencies.yaml files are only used to discover upstream and downstream packages. These are called internal packages. Then, the true dependency graph based on the DESCRIPTION files is constructed. All listed upstream dependencies that are not internal are called external dependencies. There exists a function to check that the staged_dependencies.yaml files are consistent: if package x lists y as a downstream package, then y should list x as an upstream package.
When testing the addins, note that they run in a separate R process, so they pick up the currently installed version of staged.dependencies rather than the one loaded with devtools::load_all().
Troubleshooting
git2r::clone may fail. Check that the git host is reachable (VPN may be needed) and that the access token has read privileges for the repositories.
When you run remotes::install_deps for a package that was installed with this package, issues may arise because not all repositories are publicly accessible. Make sure to provide auth tokens.
Users of Macs may need to install the oskeyring package.
Troubleshooting (For Developers of This Package)
For developing this package, we use renv. If you set environment variables such as GITHUB_PAT in your ~/.Rprofile, they will not be available because the project .Rprofile does not source the global ~/.Rprofile. To enable this, put the following into ~/.Renviron:
RENV_CONFIG_USER_PROFILE=TRUEMake sure that ~/.Rprofile does not set lib paths as this may interfere with renv. Also see the renv doc.
Example Setup
This package can also be tested on a more complex setup of packages available at: https://github.com/openpharma/stageddeps.elecinfra https://github.com/openpharma/stageddeps.electricity https://github.com/openpharma/stageddeps.food https://github.com/openpharma/stageddeps.garden https://github.com/openpharma/stageddeps.house https://github.com/openpharma/stageddeps.water
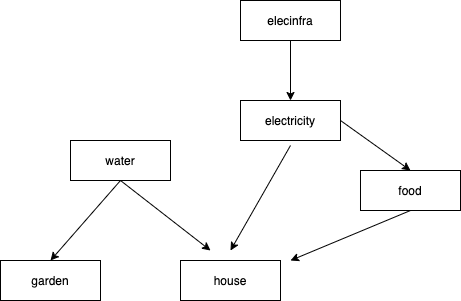
You can check that the right branches of the packages are installed with:
stageddeps.house::get_house()
stageddeps.water::get_water()
stageddeps.garden::get_garden()
stageddeps.food::get_food()
stageddeps.elecinfra::get_elecinfra()
stageddeps.electricity::get_electricity()