Common multiple comparison procedures illustrated using graphicalMCP
Source:vignettes/graph-examples.Rmd
graph-examples.RmdIntroduction
In confirmatory clinical trials, regulatory guidelines mandate the strong control of the family-wise error rate at a prespecified level . Many multiple comparison procedures (MCPs) have been proposed for this purpose. The graphical approaches are a general framework that include many common MCPs as special cases. In this vignette, we illustrate how to use graphicalMCP to perform some common MCPs.
To test hypotheses using a graphical MCP, each hypothesis receives a weight (called hypothesis weight), where . From to , there could be a directed and weighted edge , which means that when is rejected, its hypothesis weight will be propagated (or transferred) to and determines how much of the propagation. We also require and .
Bonferroni-based procedures
Bonferroni test
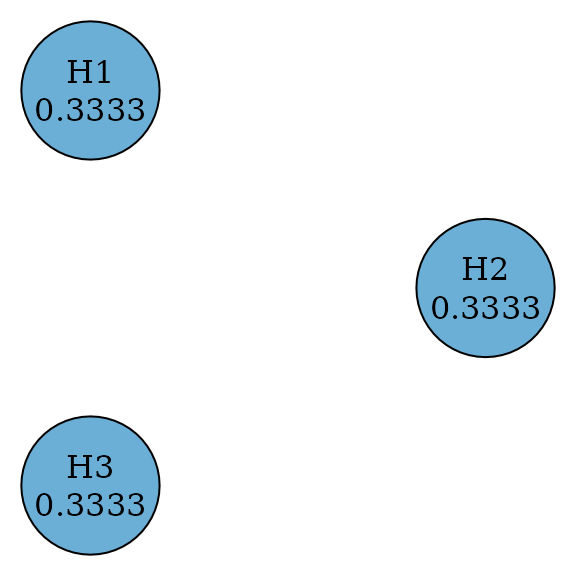
A Bonferroni test splits equally among hypotheses by testing every hypothesis at a significance level of divided by the number of hypotheses. Thus it rejects a hypothesis if its p-value is less than or equal to its significance level. There is no propagation between any pair of hypothesis.
set.seed(1234)
alpha <- 0.025
m <- 3
bonferroni_graph <- bonferroni(m)
# transitions <- matrix(0, m, m)
# bonferroni_graph <- graph_create(rep(1 / m, m), transitions)
plot(
bonferroni_graph,
layout = igraph::layout_in_circle(
as_igraph(bonferroni_graph),
order = c(2, 1, 3)
),
vertex.size = 70
)
p_values <- runif(m, 0, alpha)
test_results <-
graph_test_shortcut(
bonferroni_graph,
p = p_values,
alpha = alpha
)
test_results$outputs$rejected
#> H1 H2 H3
#> TRUE FALSE FALSEWeighted Bonferroni test
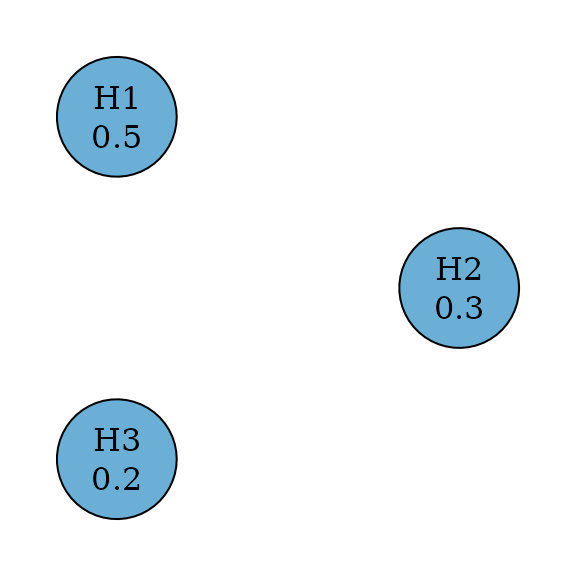
A weighted Bonferroni test splits among hypotheses by testing every hypothesis at a significance level of . Thus it rejects a hypothesis if its p-value is less than or equal to its significance level. When for all , this means an equal split and the test is the Bonferroni test. There is no propagation between any pair of hypothesis.
set.seed(1234)
alpha <- 0.025
hypotheses <- c(0.5, 0.3, 0.2)
weighted_bonferroni_graph <- bonferroni_weighted(hypotheses)
# m <- length(hypotheses)
# transitions <- matrix(0, m, m)
# weighted_bonferroni_graph <- graph_create(hypotheses, transitions)
plot(
weighted_bonferroni_graph,
layout = igraph::layout_in_circle(
as_igraph(bonferroni_graph),
order = c(2, 1, 3)
),
vertex.size = 70
)
p_values <- runif(m, 0, alpha)
test_results <-
graph_test_shortcut(
weighted_bonferroni_graph,
p = p_values,
alpha = alpha
)
test_results$outputs$rejected
#> H1 H2 H3
#> TRUE FALSE FALSEHolm Procedure
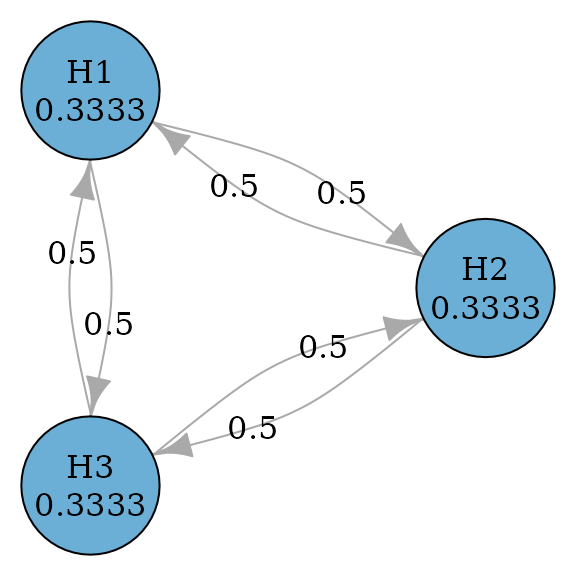
Holm (or Bonferroni-Holm) procedures improve over Bonferroni tests by allowing propagation (Holm 1979). In other words, transition weights between hypotheses may not be zero. So it is uniformly more powerful than Bonferroni tests.
set.seed(1234)
alpha <- 0.025
m <- 3
holm_graph <- bonferroni_holm(m)
# transitions <- matrix(1 / (m - 1), m, m)
# diag(transitions) <- 0
# holm_graph <- graph_create(rep(1 / m, m), transitions)
plot(holm_graph,
layout = igraph::layout_in_circle(
as_igraph(holm_graph),
order = c(2, 1, 3)
),
vertex.size = 70
)
p_values <- runif(m, 0, alpha)
test_results <- graph_test_shortcut(holm_graph, p = p_values, alpha = alpha)
test_results$outputs$rejected
#> H1 H2 H3
#> TRUE FALSE FALSEWeighted Holm Procedure
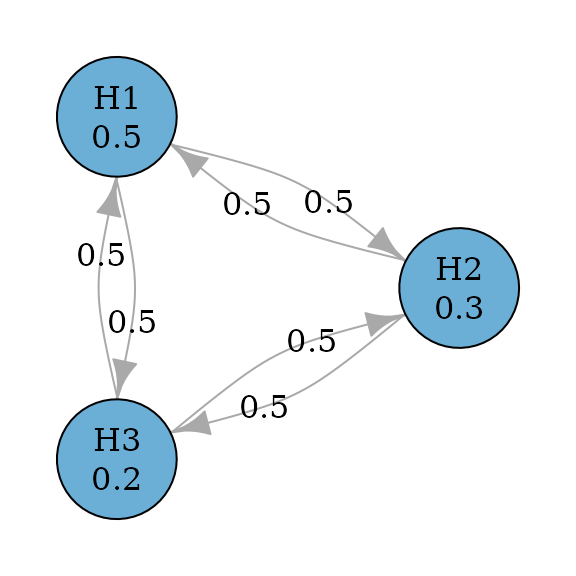
Weighted Holm (or weighted Bonferroni-Holm) procedures improve over weighted Bonferroni tests by allowing propagation (Holm 1979). In other words, transition weights between hypotheses may not be zero. So it is uniformly more powerful than weighted Bonferroni tests.
set.seed(1234)
alpha <- 0.025
hypotheses <- c(0.5, 0.3, 0.2)
weighted_holm_graph <- bonferroni_holm_weighted(hypotheses)
# m <- length(hypotheses)
# transitions <- matrix(1 / (m - 1), m, m)
# diag(transitions) <- 0
# weighted_holm_graph <- graph_create(hypotheses, transitions)
plot(weighted_holm_graph,
layout = igraph::layout_in_circle(
as_igraph(holm_graph),
order = c(2, 1, 3)
),
vertex.size = 70
)
p_values <- runif(m, 0, alpha)
test_results <- graph_test_shortcut(weighted_holm_graph, p = p_values, alpha = alpha)
test_results$outputs$rejected
#> H1 H2 H3
#> TRUE FALSE FALSEFixed sequence procedure
Fixed sequence (or hierarchical) procedures pre-specify an order of testing Westfall and Krishen (2001). For example, the procedure will test first. If it is rejected, it will test ; otherwise the testing stops. If is rejected, it will test ; otherwise the testing stops. For each hypothesis, it will be tested at the full level, when it can be tested.
set.seed(1234)
alpha <- 0.025
m <- 3
fixed_sequence_graph <- fixed_sequence(m)
# transitions <- rbind(
# c(0, 1, 0),
# c(0, 0, 1),
# c(0, 0, 0)
# )
# fixed_sequence_graph <- graph_create(c(1, 0, 0), transitions)
plot(fixed_sequence_graph, nrow = 1, asp = 0.5, vertex.size = 50)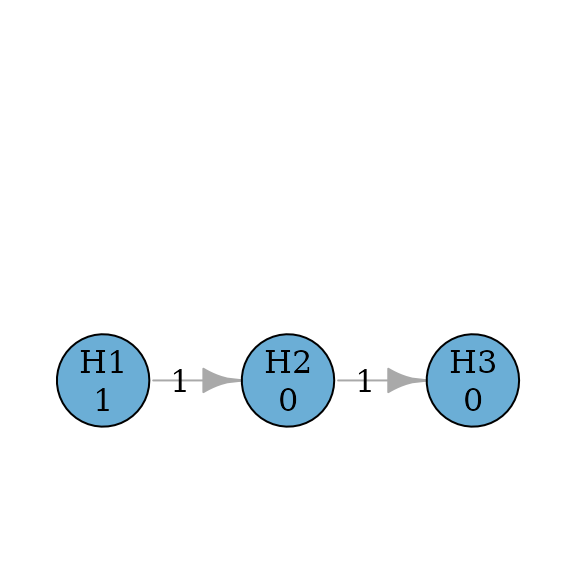
p_values <- runif(m, 0, alpha)
test_results <-
graph_test_shortcut(
fixed_sequence_graph,
p = p_values,
alpha = alpha
)
test_results$outputs$rejected
#> H1 H2 H3
#> TRUE TRUE TRUEFallback procedure
Fallback procedures have one-way propagation (like fixed sequence procedures) but allow hypotheses to be tested at different significance levels (Wiens 2003).
set.seed(1234)
alpha <- 0.025
hypotheses <- c(0.5, 0.3, 0.2)
m <- length(hypotheses)
fallback_graph <- fallback(hypotheses)
# transitions <- rbind(
# c(0, 1, 0),
# c(0, 0, 1),
# c(0, 0, 0)
# )
# fallback_graph <- graph_create(hypotheses, transitions)
plot(fallback_graph, nrow = 1, asp = 0.5, vertex.size = 50)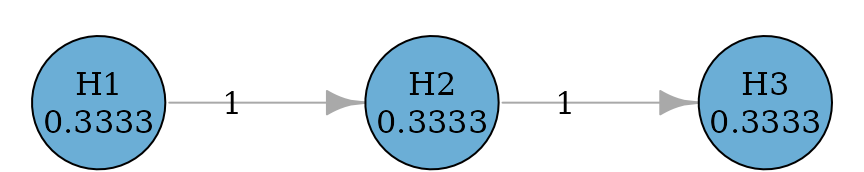
p_values <- runif(m, 0, alpha)
test_results <-
graph_test_shortcut(
fallback_graph,
p = p_values,
alpha = alpha
)
test_results$outputs$rejected
#> H1 H2 H3
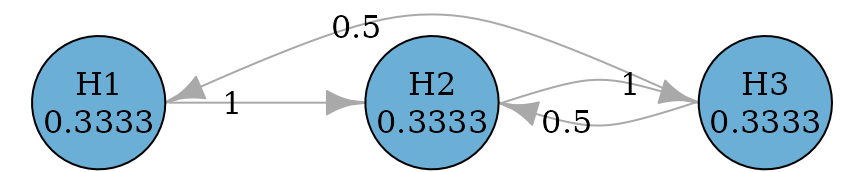
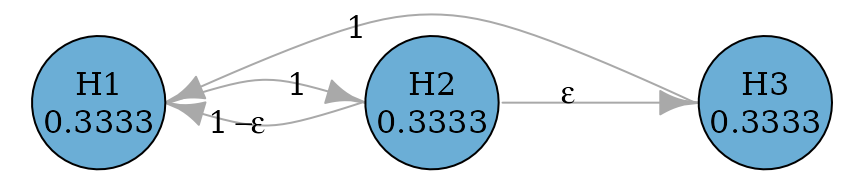
#> TRUE TRUE TRUEFurther they can be improved to allow propagation from later
hypotheses to earlier hypotheses, because it is possible that a later
hypothesis is rejected before an earlier hypothesis can be rejected.
There are two versions of improvement: fallback_improved_1
due to Wiens and Dmitrienko (2005) and
fallback_improved_2 due to Bretz et
al. (2009) respectively.
set.seed(1234)
alpha <- 0.025
hypotheses <- c(0.5, 0.3, 0.2)
m <- length(hypotheses)
fallback_improved_1_graph <- fallback_improved_1(hypotheses)
# transitions <- rbind(
# c(0, 1, 0),
# c(0, 0, 1),
# c(hypotheses[seq_len(m - 1)] / sum(hypotheses[seq_len(m - 1)]), 0)
# )
# fallback_improved_1_graph <- graph_create(hypotheses, transitions)
plot_layout <- rbind(
c(0, 0.8),
c(0.8, 0.8),
c(0.4, 0)
)
plot(
fallback_improved_1_graph,
layout = plot_layout,
asp = 0.5,
edge_curves = c(pairs = 0.5),
vertex.size = 70
)
epsilon <- 0.0001
fallback_improved_2_graph <- fallback_improved_2(hypotheses, epsilon)
# transitions <- rbind(
# c(0, 1, 0),
# c(1 - epsilon, 0, epsilon),
# c(1, 0, 0)
# )
# fallback_improved_2_graph <- graph_create(hypotheses, transitions)
plot_layout <- rbind(
c(0, 0.8),
c(0.8, 0.8),
c(0.4, 0)
)
plot(
fallback_improved_2_graph,
layout = plot_layout,
eps = 0.0001,
asp = 0.5,
edge_curves = c(pairs = 0.5),
vertex.size = 70
)
p_values <- runif(m, 0, alpha)
test_results <-
graph_test_shortcut(
fallback_improved_2_graph,
p = p_values,
alpha = alpha
)
test_results$outputs$rejected
#> H1 H2 H3
#> TRUE TRUE TRUESerial gatekeeping procedure
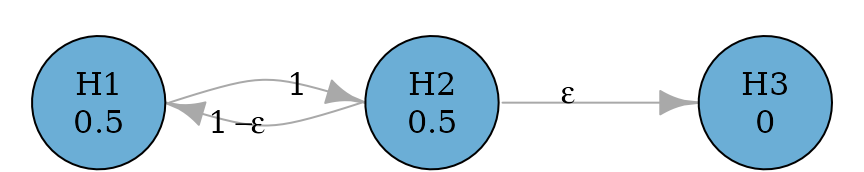
Serial gatekeeping procedures involve ordered multiple families of hypotheses, where all hypotheses of a family of hypotheses must be rejected before proceeding in the test sequence. The example below considers a primary family consisting of two hypotheses and and a secondary family consisting of a single hypothesis . In the primary family, the Holm procedure is applied. If both and are rejected, can be tested at level ; otherwise cannot be rejected. To allow the conditional propagation to , an edge is used from to . It has a very small transition weight so that propagates most of its hypothesis weight to (if not already rejected) and retains a small (non-zero) weight for so that if has been rejected, all hypothesis weight of will be propagated to . Here is assigned to be 0.0001 and in practice, the value could be adjusted but it should be much smaller than the smallest p-value observed.
set.seed(1234)
alpha <- 0.025
m <- 3
epsilon <- 0.0001
transitions <- rbind(
c(0, 1, 0),
c(1 - epsilon, 0, epsilon),
c(0, 0, 0)
)
serial_gatekeeping_graph <- graph_create(c(0.5, 0.5, 0), transitions)
plot_layout <- rbind(
c(0, 0.8),
c(0.8, 0.8),
c(0.4, 0)
)
plot(
serial_gatekeeping_graph,
layout = plot_layout,
eps = 0.0001,
asp = 0.5,
edge_curves = c(pairs = 0.5),
vertex.size = 70
)
p_values <- runif(m, 0, alpha)
test_results <-
graph_test_shortcut(
serial_gatekeeping_graph,
p = p_values,
alpha = alpha
)
test_results$outputs$rejected
#> H1 H2 H3
#> TRUE TRUE TRUEParallel gatekeeping procedure
Parallel gatekeeping procedures also involve multiple ordered families of hypotheses, where any null hypotheses of a family of hypotheses must be rejected before proceeding in the test sequence (Dmitrienko, Offen, and Westfall 2003). The example below considers a primary family consisting of two hypotheses and and a secondary family consisting of two hypotheses and . In the primary family, the Bonferroni test is applied. If any of and is rejected, and can be tested at level using the Holm procedure; if both and are rejected, and can be tested at level using the Holm procedure; otherwise and cannot be rejected.
set.seed(1234)
alpha <- 0.025
m <- 4
transitions <- rbind(
c(0, 0, 0.5, 0.5),
c(0, 0, 0.5, 0.5),
c(0, 0, 0, 1),
c(0, 0, 1, 0)
)
parallel_gatekeeping_graph <- graph_create(c(0.5, 0.5, 0, 0), transitions)
plot(parallel_gatekeeping_graph, vertex.size = 70)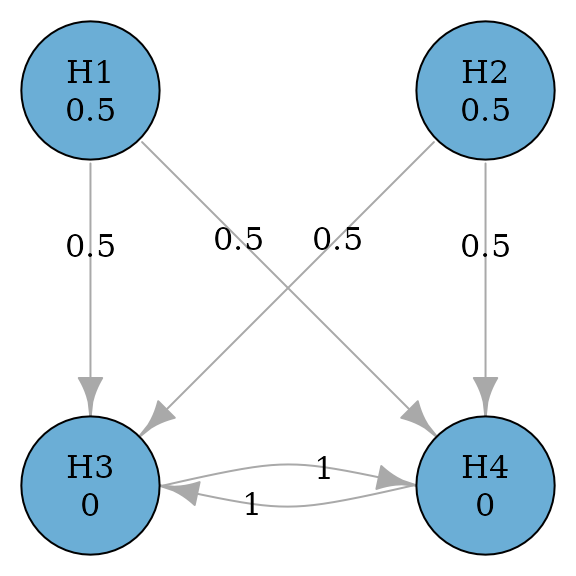
p_values <- runif(m, 0, alpha)
test_results <-
graph_test_shortcut(
parallel_gatekeeping_graph,
p = p_values,
alpha = alpha
)
test_results$outputs$rejected
#> H1 H2 H3 H4
#> TRUE FALSE FALSE FALSEThe above parallel gatekeeping procedure can be improved by adding edges from secondary hypotheses to primary hypotheses, because it is possible that both secondary hypotheses are rejected but there is still a remaining primary hypothesis not rejected (Bretz et al. 2009).
set.seed(1234)
alpha <- 0.025
m <- 4
epsilon <- 0.0001
transitions <- rbind(
c(0, 0, 0.5, 0.5),
c(0, 0, 0.5, 0.5),
c(epsilon, 0, 0, 1 - epsilon),
c(0, epsilon, 1 - epsilon, 0)
)
parallel_gatekeeping_improved_graph <-
graph_create(c(0.5, 0.5, 0, 0), transitions)
plot(parallel_gatekeeping_improved_graph, eps = 0.0001, vertex.size = 70)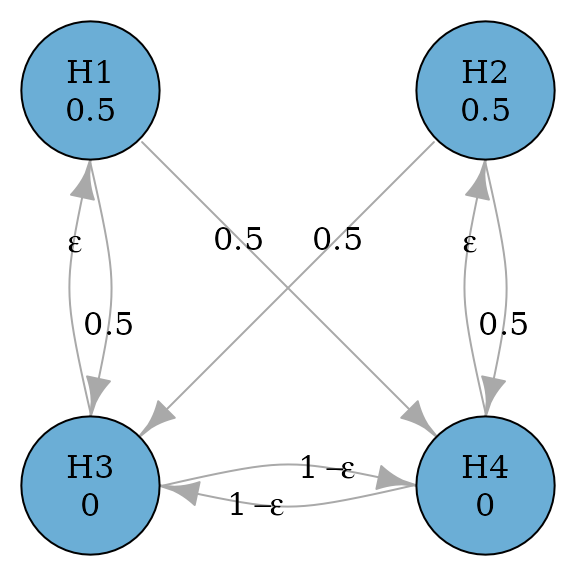
p_values <- runif(m, 0, alpha)
test_results <-
graph_test_shortcut(
parallel_gatekeeping_improved_graph,
p = p_values,
alpha = alpha
)
test_results$outputs$rejected
#> H1 H2 H3 H4
#> TRUE FALSE FALSE FALSESuccessive procedure
Successive procedures incorporate successive relationships between hypotheses. For example, the secondary hypothesis is not tested until the primary hypothesis has been rejected. This is similar to using the fixed sequence procedure as a component of a graph. The example below considers two primary hypotheses and and two secondary hypotheses and . Primary hypotheses and receive the equal hypothesis weight of 0.5; secondary hypotheses and receive the hypothesis weight of 0. A secondary hypothesis can be tested only if the corresponding primary hypothesis has been rejected. This represents the successive relationships between and , and and , respectively (Maurer, Glimm, and Bretz 2011). If both and are rejected, their hypothesis weights are propagated to and , and vice versa.
set.seed(1234)
alpha <- 0.025
m <- 4
simple_successive_graph <- simple_successive_1()
# transitions <- rbind(
# c(0, 0, 1, 0),
# c(0, 0, 0, 1),
# c(0, 1, 0, 0),
# c(1, 0, 0, 0)
# )
# simple_successive_graph <- graph_create(c(0.5, 0.5, 0, 0), transitions)
plot(simple_successive_graph, layout = "grid", nrow = 2, vertex.size = 70)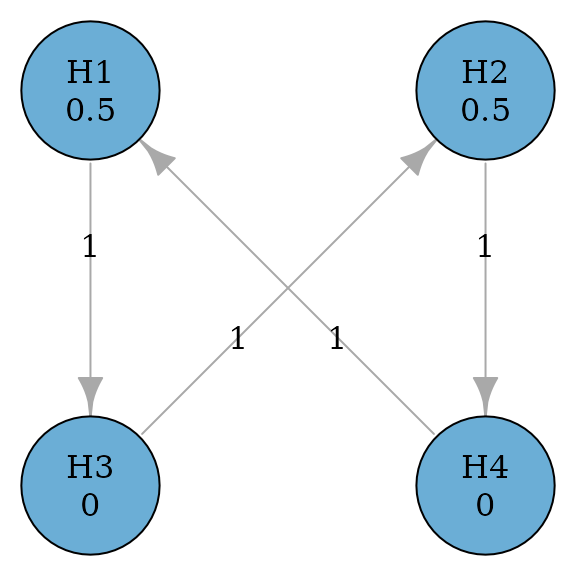
p_values <- runif(m, 0, alpha)
test_results <-
graph_test_shortcut(
simple_successive_graph,
p = p_values,
alpha = alpha
)
test_results$outputs$rejected
#> H1 H2 H3 H4
#> TRUE FALSE FALSE FALSEThe above graph could be generalized to allow propagation between primary hypotheses (Maurer, Glimm, and Bretz 2011). A general successive graph is illustrate below with a variable to determine the propagation between and .
set.seed(1234)
alpha <- 0.025
m <- 4
successive_var <- simple_successive_var <- function(gamma) {
graph_create(
c(0.5, 0.5, 0, 0),
rbind(
c(0, gamma, 1 - gamma, 0),
c(gamma, 0, 0, 1 - gamma),
c(0, 1, 0, 0),
c(1, 0, 0, 0)
)
)
}
successive_var_graph <- successive_var(0.5)
plot(successive_var_graph, layout = "grid", nrow = 2, vertex.size = 70)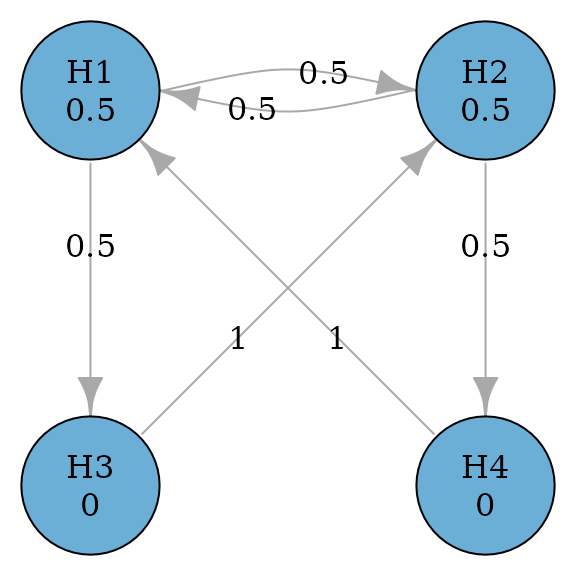
p_values <- runif(m, 0, alpha)
test_results <-
graph_test_shortcut(
successive_var_graph,
p = p_values,
alpha = alpha
)
test_results$outputs$rejected
#> H1 H2 H3 H4
#> TRUE TRUE FALSE FALSEHochberg-based procedures
Hochberg procedure
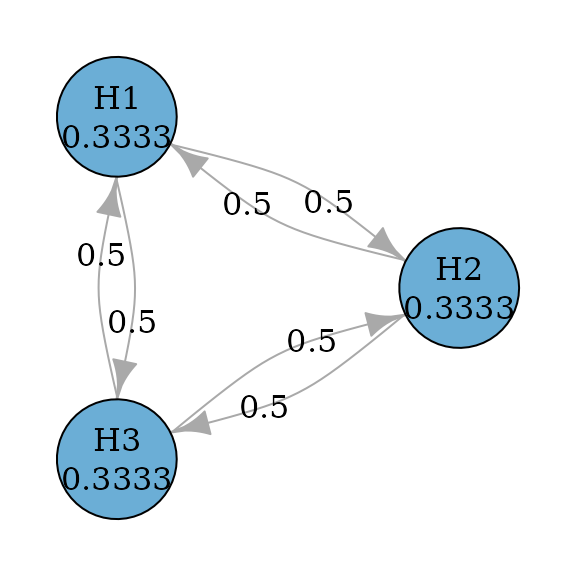
Hochberg procedure (Hochberg 1988) is a
closed test procedure which uses Hochberg tests for every intersection
hypothesis. According to Xi and Bretz
(2019), the graph for Hochberg procedures is the same as the
graph for Holm procedures. Thus to perform Hochberg procedure, we just
need to specify test_type to be hochberg.
set.seed(1234)
alpha <- 0.025
m <- 3
hochberg_graph <- hochberg(m)
plot(
hochberg_graph,
layout = igraph::layout_in_circle(
as_igraph(hochberg_graph),
order = c(2, 1, 3)
),
vertex.size = 70
)
p_values <- runif(m, 0, alpha)
test_results <- graph_test_closure(
hochberg_graph,
p = p_values,
alpha = alpha,
test_types = "hochberg"
)
test_results$outputs$rejected
#> H1 H2 H3
#> TRUE TRUE TRUESimes-based procedures
Hommel procedure
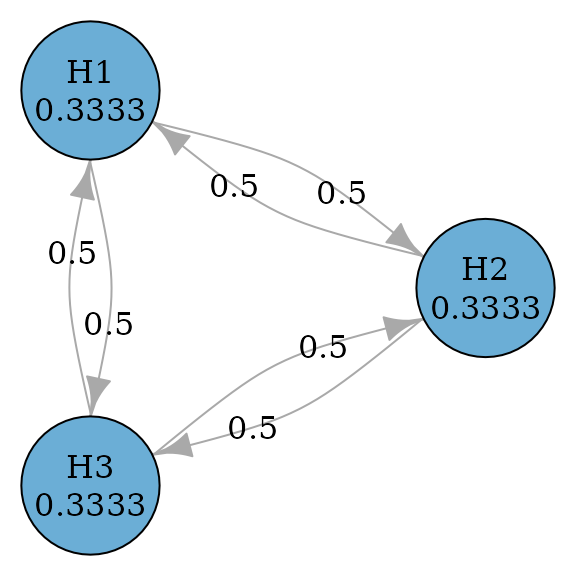
Hommel procedure (Hommel 1988) is a
closed test procedure which uses Simes tests for every intersection
hypothesis. According to Xi and Bretz
(2019), the graph for Hommel procedures is the same as the graph
for Holm procedures. Thus to perform Hommel procedure, we just need to
specify test_type to be simes.
set.seed(1234)
alpha <- 0.025
m <- 3
hommel_graph <- hommel(m)
plot(
hommel_graph,
layout = igraph::layout_in_circle(
as_igraph(hommel_graph),
order = c(2, 1, 3)
),
vertex.size = 70
)
p_values <- runif(m, 0, alpha)
test_results <- graph_test_closure(
hommel_graph,
p = p_values,
alpha = alpha,
test_types = "simes"
)
test_results$outputs$rejected
#> H1 H2 H3
#> TRUE TRUE TRUEParametric procedures
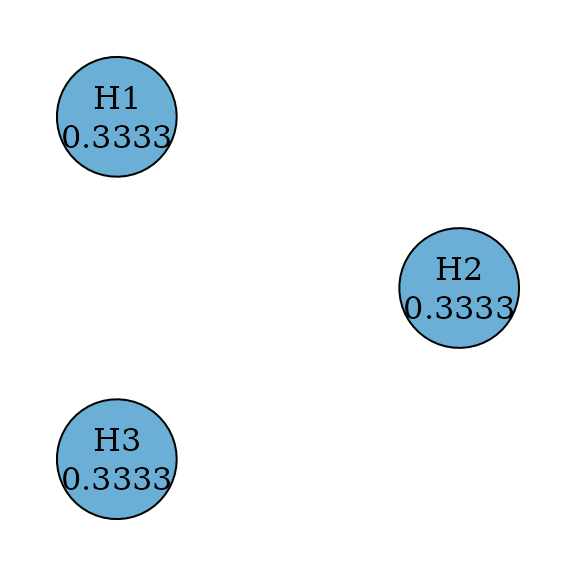
Šidák test
The Šidák test is similar to the equally weighted Bonferroni test but it assumes test statistics are independent of each other (Šidák 1967). Thus it can be performed as a parametric procedure with the identity correlation matrix. Its graph is the same as the graph for the equally weighted Bonferroni test.
set.seed(1234)
alpha <- 0.025
m <- 3
sidak_graph <- sidak(m)
plot(
sidak_graph,
layout = igraph::layout_in_circle(
as_igraph(bonferroni_graph),
order = c(2, 1, 3)
),
vertex.size = 70
)
p_values <- runif(m, 0, alpha)
corr <- diag(m)
test_results <- graph_test_closure(
sidak_graph,
p = p_values, alpha = alpha,
test_types = "parametric",
test_corr = list(corr)
)
test_results$outputs$rejected
#> H1 H2 H3
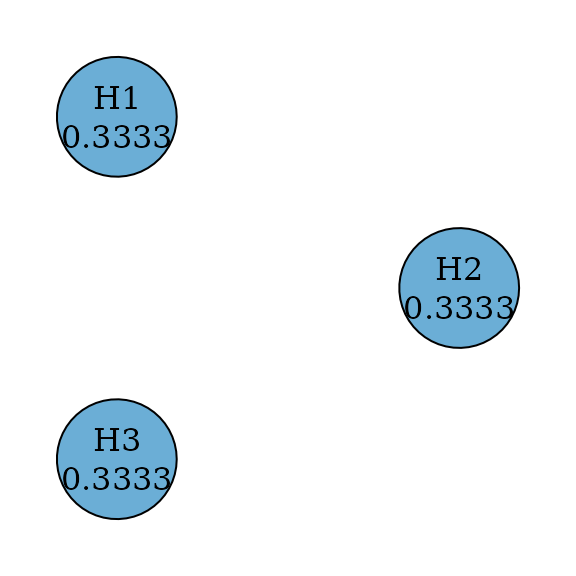
#> TRUE FALSE FALSEDunnett test
Single-step Dunnett tests are an improvement from Bonferroni tests by
incorporating the correlation structure between test statistics (Dunnett 1955). Thus their graphs are the same
as Bonferroni tests. Assume an equi-correlated case, where the
correlation between any pair of test statistics is the same, e.g., 0.5.
Then we can perform the single-step Dunnett test by specifying
test_type to be parametric and providing the
correlation matrix.
set.seed(1234)
alpha <- 0.025
m <- 3
dunnett_graph <- dunnett_single_step(m)
plot(
dunnett_graph,
layout = igraph::layout_in_circle(
as_igraph(dunnett_graph),
order = c(2, 1, 3)
),
vertex.size = 70
)
p_values <- runif(m, 0, alpha)
corr <- matrix(0.5, m, m)
diag(corr) <- 1
test_results <- graph_test_closure(
dunnett_graph,
p = p_values, alpha = alpha,
test_types = "parametric",
test_corr = list(corr)
)
test_results$outputs$rejected
#> H1 H2 H3
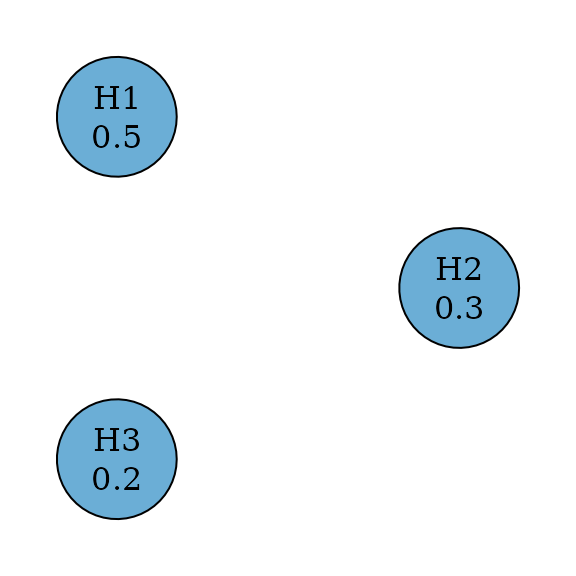
#> TRUE FALSE FALSEWeighted Dunnett test
Weighted single-step Dunnett tests are an improvement from weighted
Bonferroni tests by incorporating the correlation structure between test
statistics (Xi et al. 2017). Thus their
graphs are the same as weighted Bonferroni tests. Assume an
equi-correlated case, where the correlation between any pair of test
statistics is the same, e.g., 0.5. Then we can perform the weighted
single-step Dunnett test by specifying test_type to be
parametric and providing the correlation matrix.
set.seed(1234)
alpha <- 0.025
hypotheses <- c(0.5, 0.3, 0.2)
dunnett_graph <- dunnett_single_step_weighted(hypotheses)
plot(
dunnett_graph,
layout = igraph::layout_in_circle(
as_igraph(dunnett_graph),
order = c(2, 1, 3)
),
vertex.size = 70
)
p_values <- runif(m, 0, alpha)
corr <- matrix(0.5, m, m)
diag(corr) <- 1
test_results <- graph_test_closure(
dunnett_graph,
p = p_values, alpha = alpha,
test_types = "parametric",
test_corr = list(corr)
)
test_results$outputs$rejected
#> H1 H2 H3
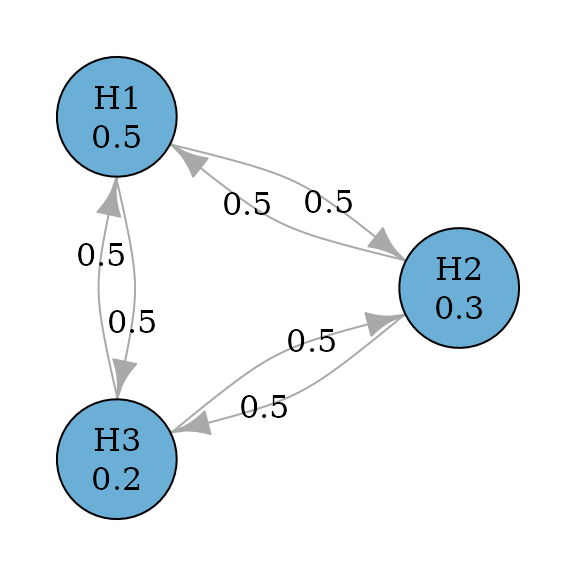
#> TRUE FALSE FALSEDunnett procedure
Dunnett procedures are a closed test procedure and an improvement
from Holm procedures by incorporating the correlation structure between
test statistics (Xi et al. 2017). Thus
their graphs are the same as Holm procedures. Assume an equi-correlated
case, where the correlation between any pair of test statistics is the
same, e.g., 0.5. Then we can perform the step-down Dunnett procedure by
specifying test_type to be parametric and
providing the correlation matrix.
set.seed(1234)
alpha <- 0.025
hypotheses <- c(0.5, 0.3, 0.2)
dunnett_graph <- dunnett_closure_weighted(hypotheses)
plot(
dunnett_graph,
layout = igraph::layout_in_circle(
as_igraph(dunnett_graph),
order = c(2, 1, 3)
),
vertex.size = 70
)
p_values <- runif(m, 0, alpha)
corr <- matrix(0.5, m, m)
diag(corr) <- 1
test_results <- graph_test_closure(
dunnett_graph,
p = p_values, alpha = alpha,
test_types = "parametric",
test_corr = list(corr)
)
test_results$outputs$rejected
#> H1 H2 H3
#> TRUE FALSE FALSE