This vignette explains how to use the pmrm package
Raw data
The package functions expect tidy longitudinal data with one row per
subject visit and columns with the patient outcome, continuous time,
indicator variables, and optional covariates. The pmrm
package has functions to simulate datasets from each supported
model.
library(pmrm)
set.seed(0)
simulation <- pmrm_simulate_decline_proportional(
patients = 500,
visit_times = c(0, 1, 2, 3, 4),
tau = 0.25,
alpha = c(0, 0.7, 1, 1.4, 1.6),
beta = c(0, 0.2, 0.35),
gamma = c(-1, 1)
)The outcome column can have missing values, but no other column must have missing values.1
simulation$y[c(2L, 3L, 12L, 13L, 14L, 15L)] <- NA_real_The output datasets includes important information that you would supply to a modeling function.
simulation[, c("y", "t", "patient", "visit", "arm")]
#> # A tibble: 2,500 × 5
#> y t patient visit arm
#> <dbl> <dbl> <chr> <ord> <ord>
#> 1 2.20 0 patient_1 visit_1 arm_1
#> 2 NA 1.47 patient_1 visit_2 arm_1
#> 3 NA 1.90 patient_1 visit_3 arm_1
#> 4 1.93 2.73 patient_1 visit_4 arm_1
#> 5 3.33 4.04 patient_1 visit_5 arm_1
#> 6 -4.75 0 patient_2 visit_1 arm_2
#> 7 0.271 1.16 patient_2 visit_2 arm_2
#> 8 1.95 1.97 patient_2 visit_3 arm_2
#> 9 1.99 3.09 patient_2 visit_4 arm_2
#> 10 3.60 4.45 patient_2 visit_5 arm_2
#> # ℹ 2,490 more rowsModels
To fit a model, simply supply the preprocessed dataset and optionally
the knot vector xi (which usually just contains the
scheduled times of all the visits). The package is powered by the RTMB package, so
the model fitting process runs fast.
system.time(
fit <- pmrm_model_decline_proportional(
data = simulation,
outcome = "y",
time = "t",
patient = "patient",
visit = "visit",
arm = "arm",
covariates = ~ w_1 + w_2,
visit_times = c(0, 1, 2, 3, 4)
)
)
#> user system elapsed
#> 0.435 0.013 0.448
print(fit)
#> Model:
#>
#> PMRM type: decline
#> Parameterization: proportional
#>
#> Fit:
#>
#> Convergence: converged
#> Observations: 2494
#> Parameters: 24
#> Log likelihood: -3555.535
#> Deviance: 7111.071
#> AIC: 7159.071
#> BIC: 7298.79
#>
#> Treatment effects:
#>
#> estimate std.error
#> arm_2 0.2493296 0.04117794
#> arm_3 0.3749138 0.04023259The fitted model object has parameter estimates, standard errors,
model constants, the convergence status, raw RTMB objects, and
other information.
names(fit)
#> [1] "data" "constants" "options" "objective"
#> [5] "model" "optimization" "report" "initial"
#> [9] "final" "estimates" "standard_errors" "metrics"
#> [13] "spline"All estimates:2
str(fit$estimates)
#> List of 9
#> $ alpha : Named num [1:5] 0.0269 0.763 0.9555 1.4059 1.6247
#> ..- attr(*, "names")= chr [1:5] "1" "2" "3" "4" ...
#> $ theta : num [1:2] 0.249 0.375
#> $ gamma : num [1:2] -0.954 0.995
#> $ phi : num [1:5] 0.0364 0.0134 -0.0022 -0.0399 0.0403
#> $ rho : num [1:10] -0.0429 -0.0824 0.0437 -0.0715 -0.0532 ...
#> $ beta : num [1:3] 0 0.249 0.375
#> $ sigma : num [1:5] 1.037 1.014 0.998 0.961 1.041
#> $ Lambda: num [1:5, 1:5] 1 -0.0428 -0.082 0.0436 -0.0712 ...
#> $ Sigma : num [1:5, 1:5] 1.0756 -0.045 -0.0849 0.0434 -0.0768 ...Just the estimates of actual parameters, omitting functions of other parameters:3
str(fit$final)
#> List of 5
#> $ alpha: Named num [1:5] 0.0269 0.763 0.9555 1.4059 1.6247
#> ..- attr(*, "names")= chr [1:5] "1" "2" "3" "4" ...
#> $ theta: num [1:2] 0.249 0.375
#> $ gamma: num [1:2] -0.954 0.995
#> $ phi : num [1:5] 0.0364 0.0134 -0.0022 -0.0399 0.0403
#> $ rho : num [1:10] -0.0429 -0.0824 0.0437 -0.0715 -0.0532 ...Checking and troubleshooting
Model-checking is an involved topic, and pmrm itself
does not go into tremendous depth. Instead, it offers only the most
basic checks on the fitted model object. The goal is to guide your
intuition and high-level understanding.
First, check the convergence code. A 0 indicates convergence, and a 1 indicates that the model diverged or a local minimum was not otherwise reached (e.g. in the case of a saddle point).
fit$optimization$convergence
#> [1] 0Next, you may wish to check that the parameter estimates and standard errors make sense.
str(fit$estimates)
#> List of 9
#> $ alpha : Named num [1:5] 0.0269 0.763 0.9555 1.4059 1.6247
#> ..- attr(*, "names")= chr [1:5] "1" "2" "3" "4" ...
#> $ theta : num [1:2] 0.249 0.375
#> $ gamma : num [1:2] -0.954 0.995
#> $ phi : num [1:5] 0.0364 0.0134 -0.0022 -0.0399 0.0403
#> $ rho : num [1:10] -0.0429 -0.0824 0.0437 -0.0715 -0.0532 ...
#> $ beta : num [1:3] 0 0.249 0.375
#> $ sigma : num [1:5] 1.037 1.014 0.998 0.961 1.041
#> $ Lambda: num [1:5, 1:5] 1 -0.0428 -0.082 0.0436 -0.0712 ...
#> $ Sigma : num [1:5, 1:5] 1.0756 -0.045 -0.0849 0.0434 -0.0768 ...
str(fit$standard_errors)
#> List of 9
#> $ alpha : Named num [1:5] 0.0455 0.0637 0.0609 0.063 0.0636
#> ..- attr(*, "names")= chr [1:5] "1" "2" "3" "4" ...
#> $ theta : num [1:2] 0.0412 0.0402
#> $ gamma : num [1:2] 0.0206 0.0205
#> $ phi : num [1:5] 0.0316 0.0317 0.0318 0.0318 0.0318
#> $ rho : num [1:10] 0.0449 0.0451 0.0449 0.0451 0.0449 ...
#> $ beta : num [1:3] 0 0.0412 0.0402
#> $ sigma : num [1:5] 0.0328 0.0321 0.0318 0.0306 0.0331
#> $ Lambda: num [1:5, 1:5] 0 0.0447 0.0446 0.0447 0.0446 ...
#> $ Sigma : num [1:5, 1:5] 0.0681 0.0472 0.0466 0.0447 0.0486 ...Also, please check for odd behavior in the fitted spline .
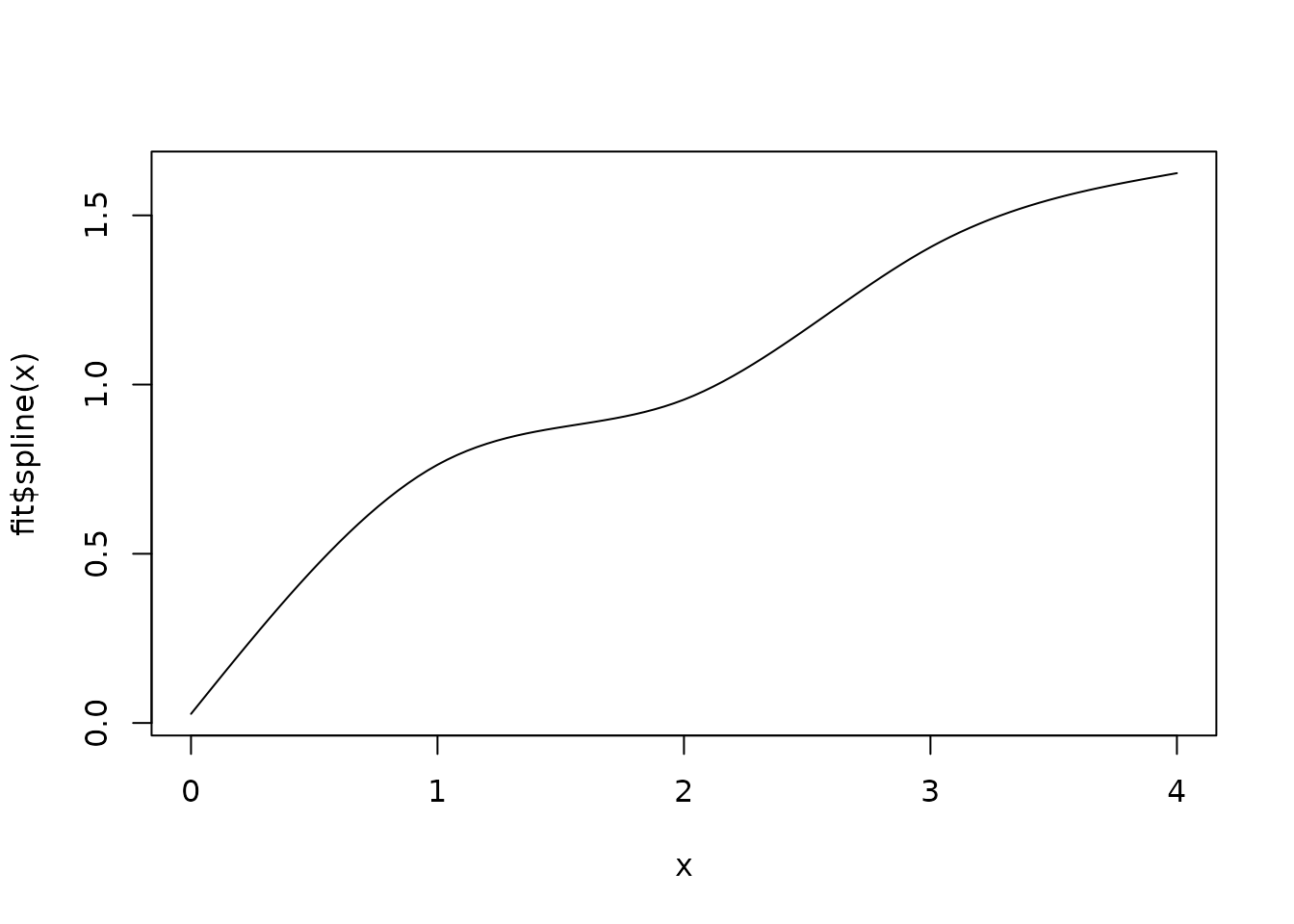
In tough cases, the spline may flatten out at the knot boundaries or
twist in strange directions. If you notice a strange fitted spline in a
divergent model, you may wish to experiment with different knot
placement or hand-pick initial values for the vertical knot distances
alpha. The latter can sometimes help stabilize
theta (especially for pmrm_model_slowing()) by
beginning with an appropriately strict monotone spline. Example:
initial <- fit$initial
initial$alpha <- c(0, 0.5, 1, 1.5, 2)
fit2 <- pmrm_model_decline_proportional(
data = simulation,
outcome = "y",
time = "t",
patient = "patient",
visit = "visit",
arm = "arm",
covariates = ~ w_1 + w_2,
visit_times = c(0, 1, 2, 3, 4),
initial = initial # with hand-picked initial alpha
)Alternatively, you can even tweak one or more parameters and then resume the original optimization.
initial <- fit$final # true parameters from the last fit
initial$theta[2] <- 0 # reset a scalar parameter that may have diverged
fit3 <- pmrm_model_decline_proportional(
data = simulation,
outcome = "y",
time = "t",
patient = "patient",
visit = "visit",
arm = "arm",
covariates = ~ w_1 + w_2,
visit_times = c(0, 1, 2, 3, 4),
initial = initial # with the modified parameter values
)Finally, you may wish to fit a Bayesian version the model using
tmbstan and check for correlations among the parameters.
Strong pairwise correlations involving the
parameter may indicate that your spline has too many knots.
library(dplyr)
library(tmbstan)
library(posterior)
# Fit a Bayesian version of the same model.
# Increase the number of chains for the Rhat diagnostic to be valid.
mcmc <- tmbstan(fit$model, chains = 1, refresh = 10)
# Extract the MCMC draws.
draws <- as_draws_df(mcmc) |>
select(starts_with("alpha"), any_of(c("theta", "phi")))
# Plot pairwise correlations among MCMC draws.
# Pairs of parameters should not be strongly correlated.
# Tight correlations and funneling indicate problems,
# such as non-identifiability if you have too many knots.
pairs(draws)
# Alternatively, if you have many parameters, you can look at
# pairwise linear correlations. However, this may miss
# important pathologies.
correlations <- cor(samples)[lower.tri(cor(samples))]
hist(correlations)Summaries
Use pmrm_estimates() to compute estimates, standard
errors, and confidence intervals of model parameters and standard
downstream functions of parameters. For example, here are the active-arm
post-baseline treatment effect parameters:
pmrm_estimates(fit, parameter = "theta")
#> # A tibble: 2 × 6
#> parameter arm estimate standard_error lower upper
#> <chr> <ord> <dbl> <dbl> <dbl> <dbl>
#> 1 theta arm_2 0.249 0.0412 0.169 0.330
#> 2 theta arm_3 0.375 0.0402 0.296 0.454And the same for the visit-specific standard deviations of the residuals:
pmrm_estimates(fit, parameter = "sigma", confidence = 0.9)
#> # A tibble: 5 × 6
#> parameter visit estimate standard_error lower upper
#> <chr> <ord> <dbl> <dbl> <dbl> <dbl>
#> 1 sigma visit_1 1.04 0.0328 0.983 1.09
#> 2 sigma visit_2 1.01 0.0321 0.961 1.07
#> 3 sigma visit_3 0.998 0.0318 0.946 1.05
#> 4 sigma visit_4 0.961 0.0306 0.911 1.01
#> 5 sigma visit_5 1.04 0.0331 0.987 1.10The summary() method produces high-level metrics for
model comparison.
summary(fit)
#> # A tibble: 1 × 8
#> model parameterization n_observations n_parameters log_likelihood deviance
#> <chr> <chr> <int> <int> <dbl> <dbl>
#> 1 decline proportional 2494 24 -3556. 7111.
#> # ℹ 2 more variables: aic <dbl>, bic <dbl>Marginals
pmrm_marginals() returns estimated marginal means for
each study arm and visit. The continuous time at each visit is given by
the marginals argument to
pmrm_model_decline_proportional().
pmrm_marginals(fit, type = "outcome")
#> # A tibble: 15 × 7
#> arm visit time estimate standard_error lower upper
#> <ord> <ord> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 arm_1 visit_1 0 0.0269 0.0455 -0.0623 0.116
#> 2 arm_1 visit_2 1 0.763 0.0637 0.638 0.888
#> 3 arm_1 visit_3 2 0.955 0.0609 0.836 1.07
#> 4 arm_1 visit_4 3 1.41 0.0630 1.28 1.53
#> 5 arm_1 visit_5 4 1.62 0.0636 1.50 1.75
#> 6 arm_2 visit_1 0 0.0269 0.0455 -0.0623 0.116
#> 7 arm_2 visit_2 1 0.579 0.0497 0.482 0.677
#> 8 arm_2 visit_3 2 0.724 0.0509 0.624 0.824
#> 9 arm_2 visit_4 3 1.06 0.0531 0.958 1.17
#> 10 arm_2 visit_5 4 1.23 0.0589 1.11 1.34
#> 11 arm_3 visit_1 0 0.0269 0.0455 -0.0623 0.116
#> 12 arm_3 visit_2 1 0.487 0.0432 0.402 0.572
#> 13 arm_3 visit_3 2 0.607 0.0453 0.518 0.696
#> 14 arm_3 visit_4 3 0.889 0.0511 0.789 0.989
#> 15 arm_3 visit_5 4 1.03 0.0569 0.914 1.14Select type = "change" for estimates of change from
baseline.
pmrm_marginals(fit, type = "change")
#> # A tibble: 15 × 7
#> arm visit time estimate standard_error lower upper
#> <ord> <ord> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 arm_1 visit_1 0 NA NA NA NA
#> 2 arm_1 visit_2 1 0.736 0.0825 0.574 0.898
#> 3 arm_1 visit_3 2 0.929 0.0793 0.773 1.08
#> 4 arm_1 visit_4 3 1.38 0.0757 1.23 1.53
#> 5 arm_1 visit_5 4 1.60 0.0782 1.44 1.75
#> 6 arm_2 visit_1 0 NA NA NA NA
#> 7 arm_2 visit_2 1 0.553 0.0684 0.419 0.687
#> 8 arm_2 visit_3 2 0.697 0.0696 0.561 0.833
#> 9 arm_2 visit_4 3 1.04 0.0697 0.899 1.17
#> 10 arm_2 visit_5 4 1.20 0.0770 1.05 1.35
#> 11 arm_3 visit_1 0 NA NA NA NA
#> 12 arm_3 visit_2 1 0.460 0.0612 0.340 0.580
#> 13 arm_3 visit_3 2 0.580 0.0639 0.455 0.706
#> 14 arm_3 visit_4 3 0.862 0.0685 0.728 0.996
#> 15 arm_3 visit_5 4 0.999 0.0762 0.849 1.15Select type = "effect" for estimates of the treatment
effect (change from baseline of each active arm minus that of the
control arm).
pmrm_marginals(fit, type = "effect")
#> # A tibble: 15 × 7
#> arm visit time estimate standard_error lower upper
#> <ord> <ord> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 arm_1 visit_1 0 NA NA NA NA
#> 2 arm_1 visit_2 1 NA NA NA NA
#> 3 arm_1 visit_3 2 NA NA NA NA
#> 4 arm_1 visit_4 3 NA NA NA NA
#> 5 arm_1 visit_5 4 NA NA NA NA
#> 6 arm_2 visit_1 0 NA NA NA NA
#> 7 arm_2 visit_2 1 -0.184 0.0370 -0.256 -0.111
#> 8 arm_2 visit_3 2 -0.232 0.0437 -0.317 -0.146
#> 9 arm_2 visit_4 3 -0.344 0.0641 -0.469 -0.218
#> 10 arm_2 visit_5 4 -0.398 0.0729 -0.541 -0.255
#> 11 arm_3 visit_1 0 NA NA NA NA
#> 12 arm_3 visit_2 1 -0.276 0.0413 -0.357 -0.195
#> 13 arm_3 visit_3 2 -0.348 0.0463 -0.439 -0.257
#> 14 arm_3 visit_4 3 -0.517 0.0652 -0.645 -0.389
#> 15 arm_3 visit_5 4 -0.599 0.0736 -0.743 -0.455Predictions
The package supports a predict() method for all PMRMs.
It returns the expected values, standard errors, and confidence
intervals for the outcomes given new data. All the important columns
from the original data need to be part of the new data, except that the
outcome column can be entirely absent.
predict(fit, data = head(simulation, 5))
#> # A tibble: 5 × 8
#> patient arm visit time estimate standard_error lower upper
#> <fct> <ord> <ord> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 patient_1 arm_1 visit_1 0 0.939 0.0476 0.846 1.03
#> 2 patient_1 arm_1 visit_2 1.47 1.69 0.0521 1.59 1.80
#> 3 patient_1 arm_1 visit_3 1.90 -1.39 0.0697 -1.52 -1.25
#> 4 patient_1 arm_1 visit_4 2.73 0.622 0.0609 0.503 0.742
#> 5 patient_1 arm_1 visit_5 4.04 2.92 0.0690 2.79 3.06Plots
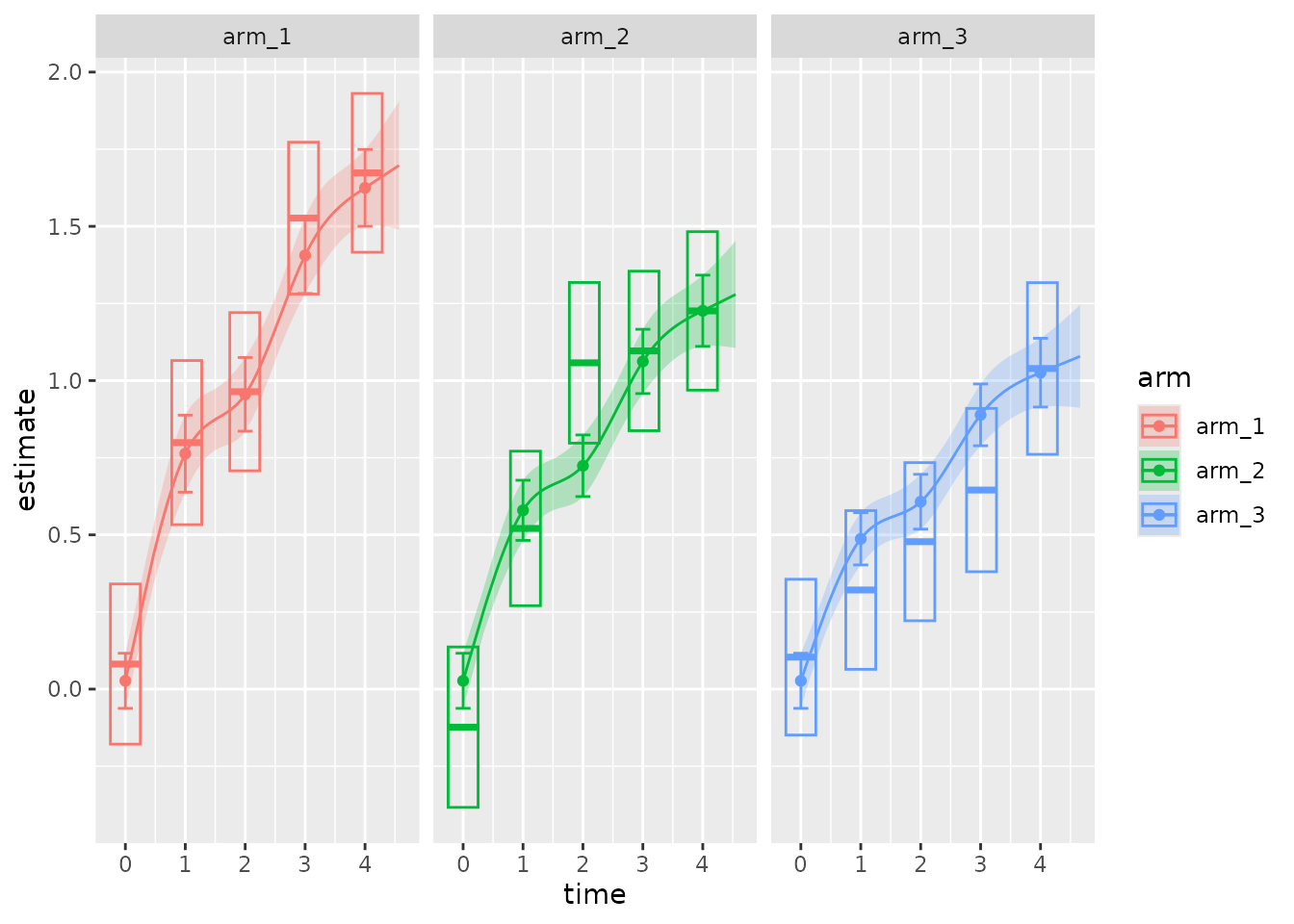
pmrm supports an S3 method for the plot()
generic. This method plot.pmrm_fit() visually compares the
model to the data. The default plot shows:
- Raw estimates and confidence intervals on the data, as points and vertical line segments.
- Model-based marginal means and confidence intervals, as horizontal line segments and boxes around them.
- Model-based predictions and confidence bands, as continuous lines and shaded regions.
plot(
fit,
show_predictions = TRUE # Defaults to FALSE because predictions take extra time.
)
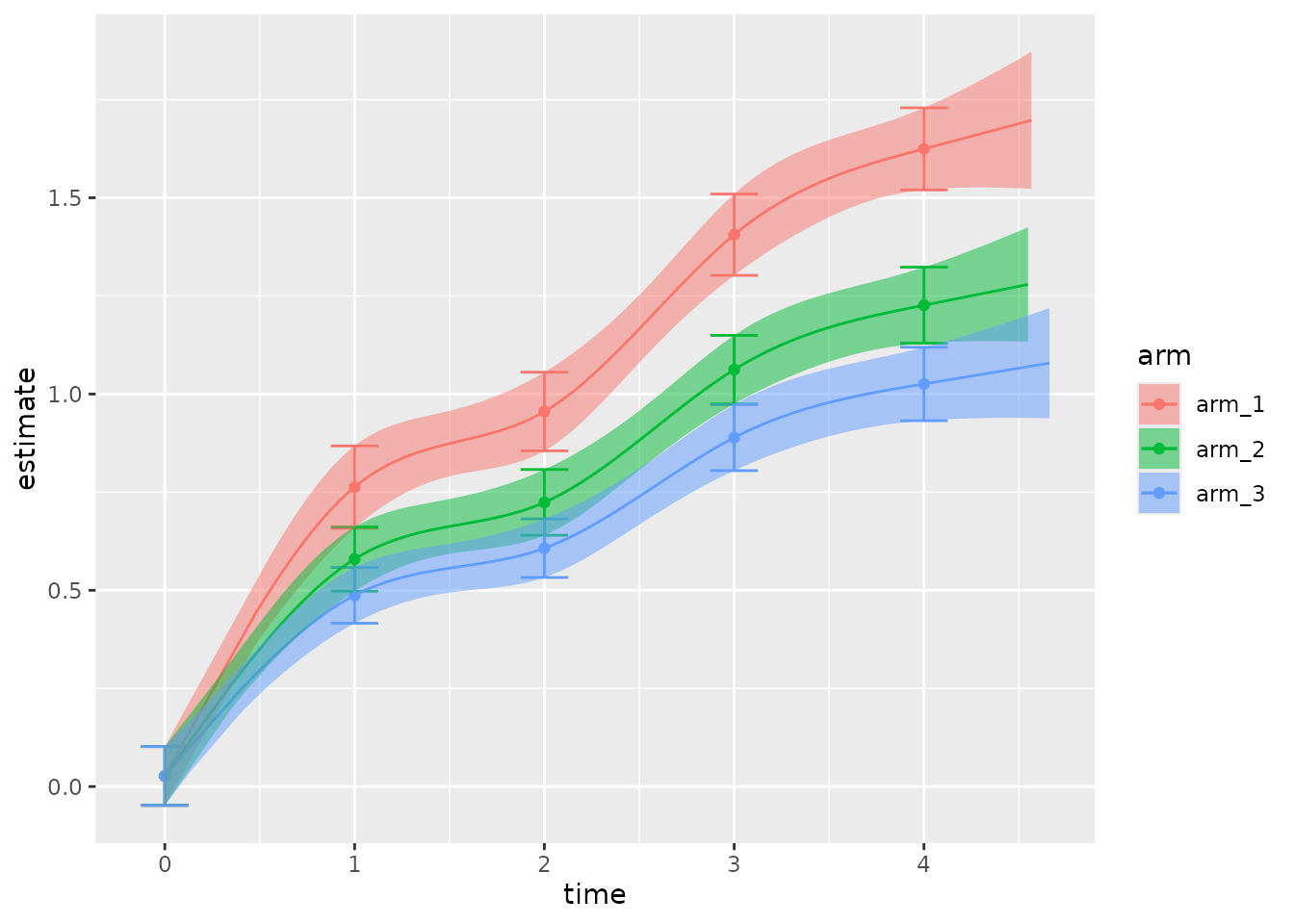
You can customize the plot: for example, to compare the fitted disease progression trajectory across treatment arms.
plot(
fit,
confidence = 0.9,
show_data = FALSE,
show_predictions = TRUE, # Defaults to FALSE because predictions take extra time.
facet = FALSE,
alpha = 0.5
)