Comparison of Bayesian and frequentist MMRM fits
08/02/2024
Source:vignettes/comparison.Rmd
comparison.RmdAbout
This vignette provides an example comparison of a Bayesian MMRM fit,
obtained by brms.mmrm::brm_model(), and a frequentist MMRM
fit, obtained by mmrm::mmrm(). An overview of parameter
estimates and differences by type of MMRM is given in the summary (Table 4) at the end.
Prerequisites
Load required packages:
> packages <- c(
+ "dplyr",
+ "tidyr",
+ "ggplot2",
+ "gt",
+ "gtsummary",
+ "purrr",
+ "parallel",
+ "brms.mmrm",
+ "mmrm",
+ "emmeans",
+ "posterior"
+ )
> invisible(lapply(packages, library, character.only = TRUE))Set seed:
> set.seed(123L)Data
Pre-processing
The fev_dat dataset is used that is contained in the
mmrm-package:
> data(fev_data, package = "mmrm")It is an artificial (simulated) dataset of a clinical trial
investigating the effect of an active treatment on FEV1
(forced expired volume in one second), compared to placebo.
FEV1 is a measure of how quickly the lungs can be emptied
and low levels may indicate chronic obstructive pulmonary disease
(COPD).
The dataset is a tibble with 800 rows and 7
variables:
-
USUBJID(subject ID), -
AVISIT(visit number), -
ARMCD(treatment,TRTorPBO), -
RACE(3-category race), -
SEX(sex), -
FEV1_BL(FEV1 at baseline, %), -
FEV1(FEV1 at study visits), -
WEIGHT(weighting variable).
In this analysis, change from baseline in FEV1 is used
as an endpoint and a corresponding new variable (FEV1_CHG)
is created.
Create and preprocess dataset for MMRM analysis:
> fev_data <- fev_data |>
+ mutate("FEV1_CHG" = FEV1 - FEV1_BL)
> fev_data <- brm_data(
+ data = fev_data,
+ outcome = "FEV1_CHG",
+ role = "change",
+ group = "ARMCD",
+ time = "AVISIT",
+ patient = "USUBJID",
+ baseline = "FEV1_BL",
+ reference_group = "PBO",
+ covariates = c("RACE", "SEX")
+ ) |>
+ mutate(ARMCD = factor(ARMCD), AVISIT = factor(AVISIT))First rows of dataset:
> head(fev_data) |>
+ gt() |>
+ tab_caption(caption = md("Table 1. First rows of dataset."))| FEV1_CHG | FEV1_BL | ARMCD | AVISIT | USUBJID | RACE | SEX |
|---|---|---|---|---|---|---|
| NA | 45.02477 | PBO | VIS1 | PT2 | Asian | Male |
| -13.569552 | 45.02477 | PBO | VIS2 | PT2 | Asian | Male |
| -8.145878 | 45.02477 | PBO | VIS3 | PT2 | Asian | Male |
| 3.783324 | 45.02477 | PBO | VIS4 | PT2 | Asian | Male |
| NA | 43.50070 | PBO | VIS1 | PT3 | Black or African American | Female |
| -7.513705 | 43.50070 | PBO | VIS2 | PT3 | Black or African American | Female |
Descriptive statistics
Table of baseline characteristics:
> fev_data |>
+ select(ARMCD, USUBJID, SEX, RACE, FEV1_BL) |>
+ distinct() |>
+ select(-USUBJID) |>
+ tbl_summary(
+ by = c(ARMCD),
+ statistic = list(
+ all_continuous() ~ "{mean} ({sd})",
+ all_categorical() ~ "{n} / {N} ({p}%)"
+ )
+ ) |>
+ modify_caption("Table 2. Baseline characteristics.")| Characteristic | PBO, N = 1051 | TRT, N = 951 |
|---|---|---|
| SEX | ||
| Male | 50 / 105 (48%) | 44 / 95 (46%) |
| Female | 55 / 105 (52%) | 51 / 95 (54%) |
| RACE | ||
| Asian | 38 / 105 (36%) | 32 / 95 (34%) |
| Black or African American | 46 / 105 (44%) | 29 / 95 (31%) |
| White | 21 / 105 (20%) | 34 / 95 (36%) |
| FEV1_BL | 40 (9) | 40 (9) |
| 1 n / N (%); Mean (SD) | ||
Table of change from baseline in FEV1 over 52 weeks:
> fev_data |>
+ pull(AVISIT) |>
+ unique() |>
+ sort() |>
+ purrr::map(
+ .f = ~ fev_data |>
+ filter(AVISIT %in% .x) |>
+ tbl_summary(
+ by = ARMCD,
+ include = FEV1_CHG,
+ type = FEV1_CHG ~ "continuous2",
+ statistic = FEV1_CHG ~ c("{mean} ({sd})",
+ "{median} ({p25}, {p75})",
+ "{min}, {max}"),
+ label = list(FEV1_CHG = paste("Visit ", .x))
+ )
+ ) |>
+ tbl_stack(quiet = TRUE) |>
+ modify_caption("Table 3. Change from baseline.")| Characteristic | PBO, N = 105 | TRT, N = 95 |
|---|---|---|
| Visit VIS1 | ||
| Mean (SD) | -8 (9) | -2 (10) |
| Median (IQR) | -9 (-16, 0) | -4 (-9, 7) |
| Range | -26, 12 | -24, 20 |
| Unknown | 37 | 29 |
| Visit VIS2 | ||
| Mean (SD) | -3 (8) | 2 (9) |
| Median (IQR) | -3 (-10, 1) | 2 (-3, 8) |
| Range | -20, 15 | -22, 23 |
| Unknown | 36 | 24 |
| Visit VIS3 | ||
| Mean (SD) | 2 (8) | 5 (9) |
| Median (IQR) | 2 (-2, 8) | 6 (0, 11) |
| Range | -15, 20 | -19, 30 |
| Unknown | 34 | 37 |
| Visit VIS4 | ||
| Mean (SD) | 8 (12) | 13 (13) |
| Median (IQR) | 6 (1, 18) | 12 (5, 22) |
| Range | -20, 39 | -14, 47 |
| Unknown | 38 | 28 |
> fev_data |>
+ group_by(ARMCD) |>
+ ggplot(aes(x = AVISIT, y = FEV1_CHG, fill = factor(ARMCD))) +
+ geom_hline(yintercept = 0, col = "grey", size = 1.2) +
+ geom_boxplot(na.rm = TRUE) +
+ labs(x = "Visit",
+ y = "Change from baseline in FEV1",
+ fill = "Treatment") +
+ scale_fill_manual(values = c("darkgoldenrod2", "coral2")) +
+ theme_bw()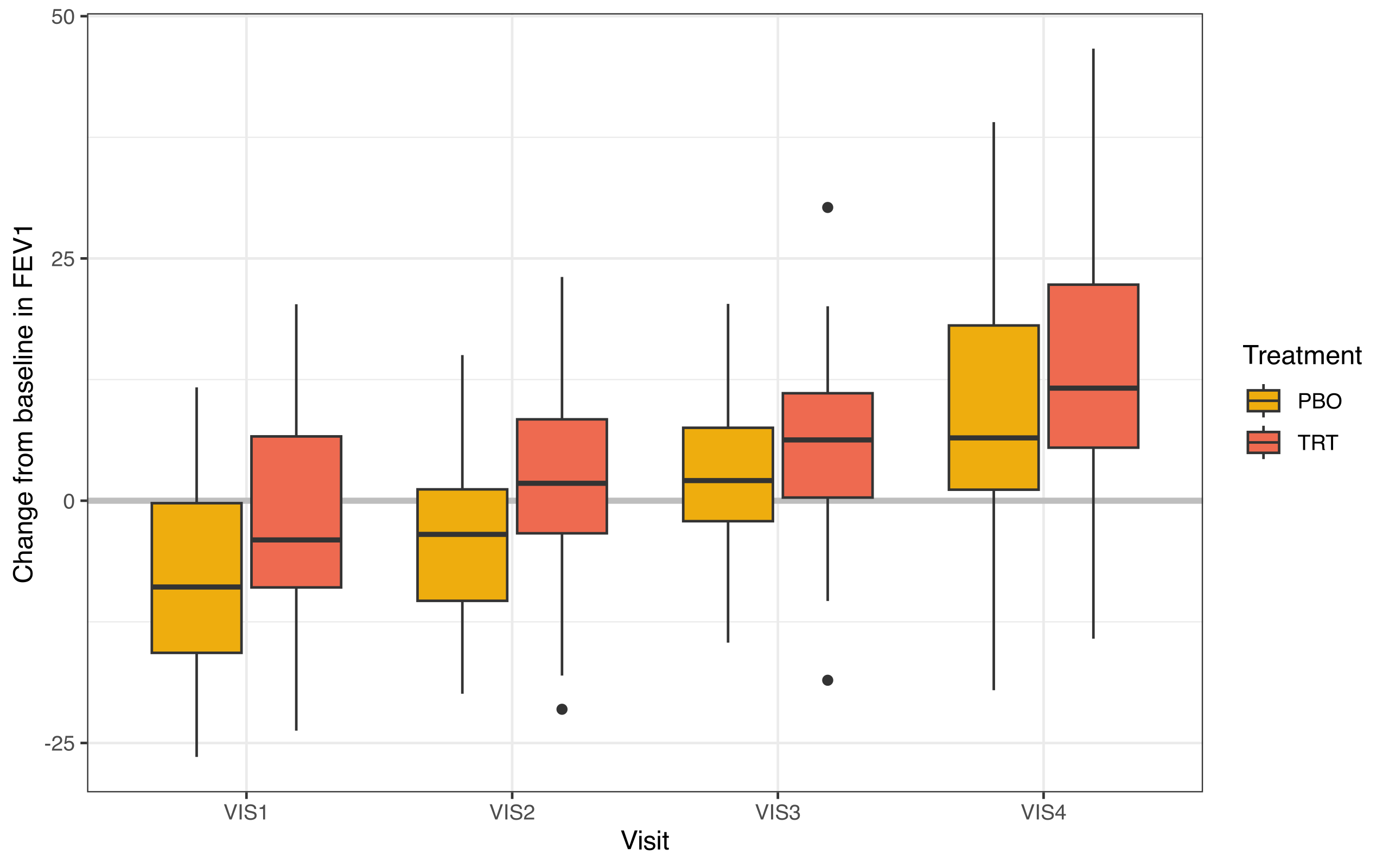
Figure 1. Change from baseline in FEV1 over 4 visit time points.
Fitting MMRMs
Bayesian model
Model formula:
> b_mmrm_formula <- brm_formula(
+ data = fev_data,
+ intercept = TRUE,
+ baseline = TRUE,
+ group = FALSE,
+ time = TRUE,
+ baseline_time = FALSE,
+ group_time = TRUE,
+ correlation = "unstructured"
+ )
> print(b_mmrm_formula)
FEV1_CHG ~ FEV1_BL + ARMCD:AVISIT + AVISIT + RACE + SEX + unstr(time = AVISIT, gr = USUBJID)
sigma ~ 0 + AVISITFit model (using multiple cores, if available, to avoid long runtime):
> n_cores <- ifelse(
+ test = detectCores() > 1,
+ yes = min(2, detectCores()),
+ no = 1
+ )> b_mmrm_fit <- brm_model(
+ data = filter(fev_data, !is.na(FEV1_CHG)),
+ formula = b_mmrm_formula,
+ iter = 1500,
+ chains = max(1, n_cores),
+ cores = n_cores,
+ refresh = 0
+ )Summary of results:
> summary(b_mmrm_fit)
Family: gaussian
Links: mu = identity; sigma = log
Formula: FEV1_CHG ~ FEV1_BL + ARMCD:AVISIT + AVISIT + RACE + SEX + unstr(time = AVISIT, gr = USUBJID)
sigma ~ 0 + AVISIT
Data: data[!is.na(data[[attr(data, "brm_outcome")]]), ] (Number of observations: 537)
Draws: 2 chains, each with iter = 1500; warmup = 750; thin = 1;
total post-warmup draws = 1500
Correlation Structures:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
cortime(VIS1,VIS2) 0.36 0.08 0.19 0.52 1.00 2550 1229
cortime(VIS1,VIS3) 0.14 0.10 -0.06 0.31 1.00 3032 1305
cortime(VIS2,VIS3) 0.04 0.10 -0.16 0.23 1.00 1928 1012
cortime(VIS1,VIS4) 0.17 0.11 -0.06 0.39 1.00 2638 1262
cortime(VIS2,VIS4) 0.11 0.09 -0.06 0.28 1.00 2404 1204
cortime(VIS3,VIS4) 0.01 0.10 -0.17 0.21 1.01 2579 855
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
Intercept 24.36 1.49 21.47 27.06 1.00 2163 1045
FEV1_BL -0.84 0.03 -0.90 -0.78 1.00 2979 1213
AVISITVIS2 4.80 0.86 3.13 6.49 1.00 1604 1119
AVISITVIS3 10.37 0.88 8.61 12.12 1.00 1338 877
AVISITVIS4 15.16 1.30 12.62 17.79 1.00 1755 1273
RACEBlackorAfricanAmerican 1.40 0.58 0.29 2.54 1.00 1983 1032
RACEWhite 5.44 0.61 4.31 6.64 1.00 2341 1238
SEXFemale 0.35 0.51 -0.63 1.36 1.00 2932 965
AVISITVIS1:ARMCDTRT 3.98 1.10 1.78 6.10 1.00 1586 960
AVISITVIS2:ARMCDTRT 3.93 0.86 2.22 5.53 1.00 2091 838
AVISITVIS3:ARMCDTRT 2.98 0.67 1.66 4.33 1.00 3024 1304
AVISITVIS4:ARMCDTRT 4.46 1.66 1.22 7.72 1.01 2110 1307
sigma_AVISITVIS1 1.83 0.06 1.72 1.96 1.00 2312 1088
sigma_AVISITVIS2 1.59 0.06 1.48 1.71 1.00 2553 1228
sigma_AVISITVIS3 1.33 0.06 1.20 1.45 1.00 2458 1188
sigma_AVISITVIS4 2.28 0.06 2.17 2.40 1.00 2565 1006
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).Frequentist model
Fit model:
> f_mmrm_fit <- mmrm::mmrm(
+ formula = FEV1_CHG ~ FEV1_BL + AVISIT + ARMCD:AVISIT + RACE + SEX +
+ us(AVISIT | USUBJID),
+ data = fev_data
+ )Summary of results:
> summary(f_mmrm_fit)
mmrm fit
Formula: FEV1_CHG ~ FEV1_BL + AVISIT + ARMCD:AVISIT + RACE + SEX + us(AVISIT | USUBJID)
Data: fev_data (used 537 observations from 197 subjects with maximum 4 timepoints)
Covariance: unstructured (10 variance parameters)
Method: Satterthwaite
Vcov Method: Asymptotic
Inference: REML
Model selection criteria:
AIC BIC logLik deviance
3381.4 3414.2 -1680.7 3361.4
Coefficients:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 24.35372 1.40754 257.97000 17.302 < 2e-16 ***
FEV1_BL -0.84022 0.02777 190.27000 -30.251 < 2e-16 ***
AVISITVIS2 4.79036 0.79848 144.82000 5.999 1.51e-08 ***
AVISITVIS3 10.36601 0.81318 157.08000 12.748 < 2e-16 ***
AVISITVIS4 15.19231 1.30857 139.25000 11.610 < 2e-16 ***
RACEBlack or African American 1.41921 0.57874 169.56000 2.452 0.015211 *
RACEWhite 5.45679 0.61626 157.54000 8.855 1.65e-15 ***
SEXFemale 0.33812 0.49273 166.43000 0.686 0.493529
AVISITVIS1:ARMCDTRT 3.98329 1.04540 142.32000 3.810 0.000206 ***
AVISITVIS2:ARMCDTRT 3.93076 0.81351 142.26000 4.832 3.46e-06 ***
AVISITVIS3:ARMCDTRT 2.98372 0.66567 129.61000 4.482 1.61e-05 ***
AVISITVIS4:ARMCDTRT 4.40400 1.66049 132.88000 2.652 0.008970 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Covariance estimate:
VIS1 VIS2 VIS3 VIS4
VIS1 37.8301 11.3255 3.4796 10.6844
VIS2 11.3255 23.5476 0.7760 5.5103
VIS3 3.4796 0.7760 13.8037 0.5683
VIS4 10.6844 5.5103 0.5683 92.9625Comparison
Extract estimates from Bayesian model
> b_mmrm_draws <- b_mmrm_fit |> as_draws_df()
> visit_levels <- sort(unique(as.character(fev_data$AVISIT)))
> for (level in visit_levels) {
+ name <- paste0("b_sigma_AVISIT", level)
+ b_mmrm_draws[[name]] <- exp(b_mmrm_draws[[name]])
+ }
> b_mmrm_summary <- b_mmrm_draws |>
+ summarize_draws() |>
+ select(variable, mean, sd) |>
+ filter(!(variable %in% c("lprior", "lp__"))) |>
+ rename(bayes_estimate = mean, bayes_se = sd) |>
+ mutate(
+ variable = variable |>
+ tolower() |>
+ gsub(pattern = "b_", replacement = "") |>
+ gsub(pattern = "b_sigma_AVISIT", replacement = "sigma_") |>
+ gsub(pattern = "cortime", replacement = "correlation") |>
+ gsub(pattern = "__", replacement = "_")
+ )Extract estimates from frequentist model
> f_mmrm_fixed <- summary(f_mmrm_fit)$coefficients |>
+ as_tibble(rownames = "variable") |>
+ mutate(variable = tolower(variable)) |>
+ mutate(variable = gsub("(", "", variable, fixed = TRUE)) |>
+ mutate(variable = gsub(")", "", variable, fixed = TRUE)) |>
+ rename(freq_estimate = Estimate, freq_se = `Std. Error`) |>
+ select(variable, freq_estimate, freq_se)> f_mmrm_variance <- tibble(
+ variable = paste0("sigma_AVISIT", visit_levels) |> tolower(),
+ freq_estimate = sqrt(diag(f_mmrm_fit$cov))
+ )> f_diagonal_factor <- diag(1 / sqrt(diag(f_mmrm_fit$cov)))
> f_corr_matrix <- f_diagonal_factor %*% f_mmrm_fit$cov %*% f_diagonal_factor
> colnames(f_corr_matrix) <- visit_levels> f_mmrm_correlation <- f_corr_matrix |>
+ as.data.frame() |>
+ as_tibble() |>
+ mutate(x1 = visit_levels) |>
+ pivot_longer(cols = -any_of("x1"),
+ names_to = "x2",
+ values_to = "freq_estimate") |>
+ filter(as.numeric(gsub("[^0-9]", "", x1)) <
+ as.numeric(gsub("[^0-9]", "", x2))) |>
+ mutate(variable = sprintf("correlation_%s_%s", x1, x2)) |>
+ select(variable, freq_estimate)> f_mmrm_summary <- bind_rows(f_mmrm_fixed,
+ f_mmrm_variance,
+ f_mmrm_correlation) |>
+ mutate(variable = gsub("\\s+", "", variable) |> tolower())Summary
> b_f_comparison <- full_join(x = b_mmrm_summary,
+ y = f_mmrm_summary,
+ by = "variable") |>
+ mutate(diff_estimate = bayes_estimate - freq_estimate,
+ diff_se = bayes_se - freq_se) |>
+ select(variable, ends_with("estimate"), ends_with("se"))> gt(b_f_comparison) |>
+ fmt_number(decimals = 4) |>
+ tab_caption(
+ caption = md("Table 4. Contrasting Bayesian and frequentist MMRM fits.")
+ ) |>
+ tab_spanner(label = "Estimate", columns = ends_with("estimate")) |>
+ tab_spanner(label = "Standard error", columns = ends_with("se")) |>
+ cols_label(
+ variable = "Variable",
+ bayes_estimate = "Bayesian",
+ freq_estimate = "Frequentist",
+ diff_estimate = "Difference",
+ bayes_se = "Bayesian",
+ freq_se = "Frequentist",
+ diff_se = "Difference",
+ )| Variable | Estimate | Standard error | ||||
|---|---|---|---|---|---|---|
| Bayesian | Frequentist | Difference | Bayesian | Frequentist | Difference | |
| intercept | 24.3603 | 24.3537 | 0.0066 | 1.4858 | 1.4075 | 0.0783 |
| fev1_bl | −0.8403 | −0.8402 | −0.0001 | 0.0300 | 0.0278 | 0.0023 |
| avisitvis2 | 4.7974 | 4.7904 | 0.0071 | 0.8592 | 0.7985 | 0.0608 |
| avisitvis3 | 10.3694 | 10.3660 | 0.0034 | 0.8766 | 0.8132 | 0.0634 |
| avisitvis4 | 15.1647 | 15.1923 | −0.0276 | 1.2965 | 1.3086 | −0.0121 |
| raceblackorafricanamerican | 1.4031 | 1.4192 | −0.0161 | 0.5791 | 0.5787 | 0.0004 |
| racewhite | 5.4414 | 5.4568 | −0.0153 | 0.6116 | 0.6163 | −0.0047 |
| sexfemale | 0.3467 | 0.3381 | 0.0086 | 0.5137 | 0.4927 | 0.0210 |
| avisitvis1:armcdtrt | 3.9849 | 3.9833 | 0.0016 | 1.0980 | 1.0454 | 0.0526 |
| avisitvis2:armcdtrt | 3.9304 | 3.9308 | −0.0004 | 0.8612 | 0.8135 | 0.0477 |
| avisitvis3:armcdtrt | 2.9806 | 2.9837 | −0.0031 | 0.6722 | 0.6657 | 0.0065 |
| avisitvis4:armcdtrt | 4.4569 | 4.4040 | 0.0529 | 1.6552 | 1.6605 | −0.0053 |
| sigma_avisitvis1 | 6.2406 | 6.1506 | 0.0900 | 0.3997 | NA | NA |
| sigma_avisitvis2 | 4.9176 | 4.8526 | 0.0650 | 0.3011 | NA | NA |
| sigma_avisitvis3 | 3.7739 | 3.7153 | 0.0586 | 0.2386 | NA | NA |
| sigma_avisitvis4 | 9.7840 | 9.6417 | 0.1423 | 0.5906 | NA | NA |
| correlation_vis1_vis2 | 0.3617 | 0.3795 | −0.0177 | 0.0843 | NA | NA |
| correlation_vis1_vis3 | 0.1385 | 0.1523 | −0.0138 | 0.0956 | NA | NA |
| correlation_vis2_vis3 | 0.0400 | 0.0430 | −0.0031 | 0.0980 | NA | NA |
| correlation_vis1_vis4 | 0.1728 | 0.1802 | −0.0074 | 0.1138 | NA | NA |
| correlation_vis2_vis4 | 0.1123 | 0.1178 | −0.0054 | 0.0878 | NA | NA |
| correlation_vis3_vis4 | 0.0132 | 0.0159 | −0.0026 | 0.0977 | NA | NA |