Survival Analysis with visR using CDISC ADaM Time-To-Event Analysis Dataset (ADTTE)
Source:vignettes/CDISC_ADaM.Rmd
CDISC_ADaM.RmdIntroduction
This tutorial illustrates how a standard time-to-event analysis can be done very efficiently when the data set adheres to the CDISC ADaM standard. A more detailed time-to-event analysis with a more broad overview of visR’s functionality is presented in another vignette.
Global Document Setup
# Metadata Title
DATASET <- paste0("Analyis Data Time-To-Event (ADTTE)")
# Save original options()
old <- options()
# Global formatting options
options(digits = 3)
# Global ggplot settings
theme_set(theme_bw())
# Global table settings
options(DT.options = list(pageLength = 10,
language = list(search = 'Filter:'),
scrollX = TRUE))
# load ADTTE from CDISC pilot
data(adtte)
# Restore original options()
options(old)Time-to-event analysis
visR includes a wrapper function to easily display summary tables
(e.g. tableone)
# Display a summary table (e.g. tableone)
visR::tableone(adtte[,c("TRTP", "AGE")],
title = "Demographic summary" , datasource = DATASET)
#> Warning: There was 1 warning in `summarise()`.
#> ℹ In argument: `TRTP = (function (x) ...`.
#> ℹ In group 1: `all = "Total"`.
#> Caused by warning:
#> ! `fct_explicit_na()` was deprecated in forcats 1.0.0.
#> ℹ Please use `fct_na_value_to_level()` instead.
#> ℹ The deprecated feature was likely used in the visR package.
#> Please report the issue at <https://github.com/openpharma/visR/issues>.| Demographic summary | |
| Total (N=254) | |
|---|---|
| TRTP | |
| Placebo | 86 (33.9%) |
| Xanomeline High Dose | 84 (33.1%) |
| Xanomeline Low Dose | 84 (33.1%) |
| AGE | |
| Mean (SD) | 75.1 (8.25) |
| Median (IQR) | 77 (70-81) |
| Min-max | 51-89 |
| Missing | 0 (0%) |
| Data Source: Analyis Data Time-To-Event (ADTTE) | |
The wrapper function to estimate a Kaplan-Meier curve is compatible
with %>% and purrr::map functions without losing
traceability of the dataset name inside the call of the object. If a
data set adheres to the CDISC ADaM standards, only a stratifier needs to
be specified.
# Estimate a survival object
survfit_object <- adtte %>%
visR::estimate_KM(data = ., strata = "TRTP")
#> Warning: `estimate_KM()` was deprecated in visR 0.4.0.
#> ℹ Please use `ggsurvfit::ggsurvfit()` instead.
#> This warning is displayed once every 8 hours.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
#> generated.
survfit_object
#> Call: ~survival::survfit(formula = survival::Surv(AVAL, 1 - CNSR) ~
#> TRTP, data = data)
#>
#> n events median 0.95LCL 0.95UCL
#> TRTP=Placebo 86 29 NA NA NA
#> TRTP=Xanomeline High Dose 84 61 36 25 47
#> TRTP=Xanomeline Low Dose 84 62 33 28 51Given a survival object, visR includes several functions to quickly
extract additional information from the survival object (e.g. test
statistics and p-values) and a general function to display a table
(render).
# Display test statistics associated with the survival estimate
visR::render(survfit_object %>% get_pvalue(), title = "P-values", datasource = DATASET)| P-values | |||
| Equality across strata | Chisq | df | p-value |
|---|---|---|---|
| Log-Rank | 60.270 | 2.00 | <0.001 |
| Wilcoxon | 48.023 | 2.00 | <0.001 |
| Tarone-Ware | 41.850 | 2.00 | <0.001 |
| Data Source: Analyis Data Time-To-Event (ADTTE) | |||
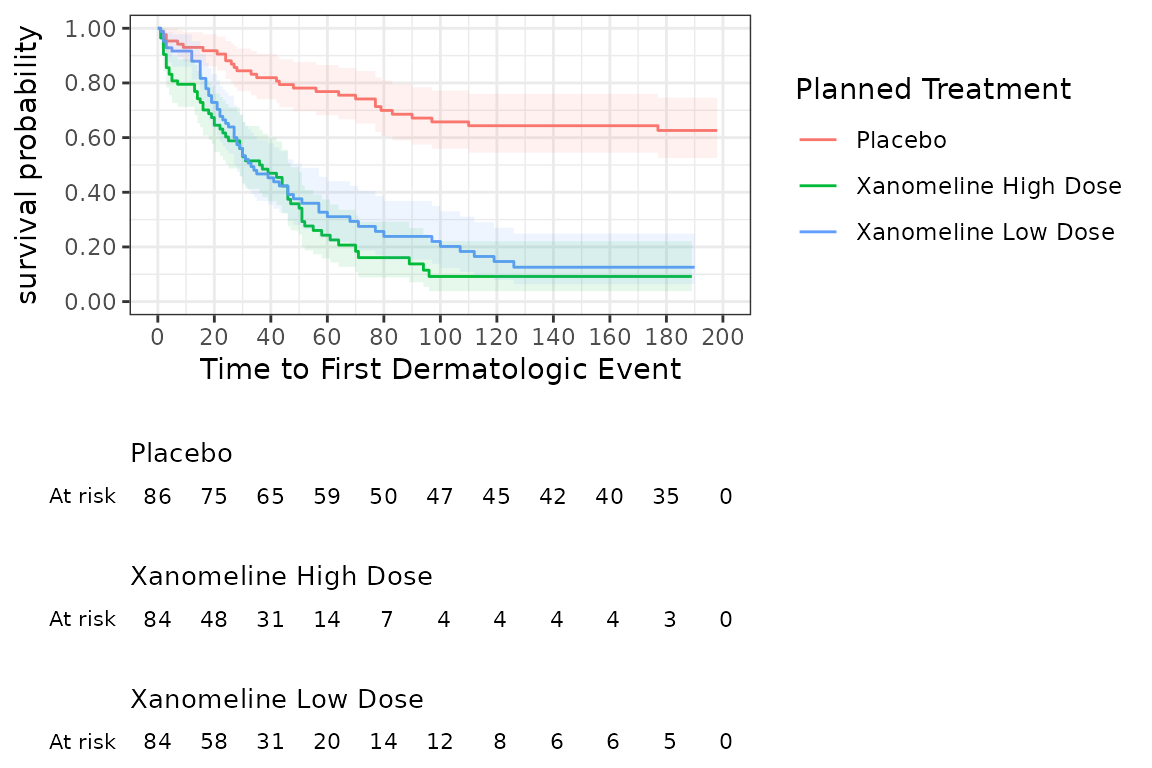
A survival object can be plotted using the visR function
visr. Additional information like confidence intervals and
a risktable can be added to the plot.
# Create and display a Kaplan-Meier from the survival object and add a risktable
visr(survfit_object) %>%
visR::add_CI() %>%
visR::add_risktable()
#> Warning: `visr.survfit()` was deprecated in visR 0.4.0.
#> ℹ Please use `ggsurvfit::ggsurvfit()` instead.
#> This warning is displayed once every 8 hours.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
#> generated.