BCVA data comparison between Bayesian and frequentist MMRMs
09/07/2024
Source:vignettes/bcva.Rmd
bcva.RmdAbout
This vignette uses the bcva_data dataset from the
mmrm package to compare a Bayesian MMRM fit, obtained by
brms.mmrm::brm_model(), and a frequentist MMRM fit,
obtained by mmrm::mmrm(). An overview of parameter
estimates and differences by type of MMRM is given in the summary (Tables 4 and 5) at the end.
Prerequisites
This comparison workflow requires the following packages.
> packages <- c(
+ "dplyr",
+ "tidyr",
+ "ggplot2",
+ "gt",
+ "gtsummary",
+ "purrr",
+ "parallel",
+ "brms.mmrm",
+ "mmrm",
+ "posterior"
+ )
> invisible(lapply(packages, library, character.only = TRUE))We set a seed for the random number generator to ensure statistical reproducibility.
Data
Pre-processing
This analysis exercise uses the bcva_data dataset
contained in the mmrm package:
According to https://openpharma.github.io/mmrm/latest-tag/articles/mmrm_review_methods.html:
The BCVA dataset contains data from a randomized longitudinal ophthalmology trial evaluating the change in baseline corrected visual acuity (BCVA) over the course of 10 visits. BCVA corresponds to the number of letters read from a visual acuity chart.
The dataset is a tibble with 8605 rows and the following
notable variables.
-
USUBJID(subject ID) -
AVISIT(visit number, factor) -
VISITN(visit number, numeric) -
ARMCD(treatment,TRTorCTL) -
RACE(3-category race) -
BCVA_BL(BCVA at baseline) -
BCVA_CHG(BCVA change from baseline, primary endpoint for the analysis)
The rest of the pre-processing steps create factors for the study arm
and visit and apply the usual checking and standardization steps of
brms.mmrm::brm_data().
> bcva_data <- bcva_data |>
+ mutate(AVISIT = gsub("VIS0*", "VIS", as.character(AVISIT))) |>
+ brm_data(
+ outcome = "BCVA_CHG",
+ group = "ARMCD",
+ time = "AVISIT",
+ patient = "USUBJID",
+ baseline = "BCVA_BL",
+ reference_group = "CTL",
+ covariates = "RACE"
+ ) |>
+ brm_data_chronologize(order = "VISITN")The following table shows the first rows of the dataset.
> head(bcva_data) |>
+ gt() |>
+ tab_caption(caption = md("Table 1. First rows of the pre-processed `bcva_data` dataset."))| USUBJID | AVISIT | VISITN | ARMCD | RACE | BCVA_BL | BCVA_CHG |
|---|---|---|---|---|---|---|
| 3 | VIS1 | 1 | CTL | Asian | 71.70881 | 5.058546 |
| 3 | VIS10 | 10 | CTL | Asian | 71.70881 | 10.152565 |
| 3 | VIS2 | 2 | CTL | Asian | 71.70881 | 4.018582 |
| 3 | VIS3 | 3 | CTL | Asian | 71.70881 | 3.572535 |
| 3 | VIS4 | 4 | CTL | Asian | 71.70881 | 4.822669 |
| 3 | VIS5 | 5 | CTL | Asian | 71.70881 | 7.348768 |
Descriptive statistics
Table of baseline characteristics:
> bcva_data |>
+ select(ARMCD, USUBJID, RACE, BCVA_BL) |>
+ distinct() |>
+ select(-USUBJID) |>
+ tbl_summary(
+ by = c(ARMCD),
+ statistic = list(
+ all_continuous() ~ "{mean} ({sd})",
+ all_categorical() ~ "{n} / {N} ({p}%)"
+ )
+ ) |>
+ modify_caption("Table 2. Baseline characteristics.")| Characteristic |
CTL N = 4941 |
TRT N = 5061 |
|---|---|---|
| RACE | ||
| Asian | 151 / 494 (31%) | 146 / 506 (29%) |
| Black | 149 / 494 (30%) | 168 / 506 (33%) |
| White | 194 / 494 (39%) | 192 / 506 (38%) |
| BCVA_BL | 75 (10) | 75 (10) |
| 1 n / N (%); Mean (SD) | ||
Table of change from baseline in BCVA over 52 weeks:
> bcva_data |>
+ pull(AVISIT) |>
+ unique() |>
+ sort() |>
+ purrr::map(
+ .f = ~ bcva_data |>
+ filter(AVISIT %in% .x) |>
+ tbl_summary(
+ by = ARMCD,
+ include = BCVA_CHG,
+ type = BCVA_CHG ~ "continuous2",
+ statistic = BCVA_CHG ~ c(
+ "{mean} ({sd})",
+ "{median} ({p25}, {p75})",
+ "{min}, {max}"
+ ),
+ label = list(BCVA_CHG = paste("Visit ", .x))
+ )
+ ) |>
+ tbl_stack(quiet = TRUE) |>
+ modify_caption("Table 3. Change from baseline.")| Characteristic |
CTL N = 494 |
TRT N = 506 |
|---|---|---|
| Visit VIS1 | ||
| Mean (SD) | 5.32 (1.23) | 5.86 (1.33) |
| Median (Q1, Q3) | 5.34 (4.51, 6.17) | 5.86 (4.98, 6.81) |
| Min, Max | 1.83, 9.02 | 2.28, 10.30 |
| Unknown | 12 | 5 |
| Visit VIS2 | ||
| Mean (SD) | 5.59 (1.49) | 6.33 (1.45) |
| Median (Q1, Q3) | 5.53 (4.64, 6.47) | 6.36 (5.34, 7.31) |
| Min, Max | 0.29, 10.15 | 2.35, 10.75 |
| Unknown | 13 | 7 |
| Visit VIS3 | ||
| Mean (SD) | 5.79 (1.61) | 6.79 (1.71) |
| Median (Q1, Q3) | 5.73 (4.64, 6.91) | 6.82 (5.66, 7.93) |
| Min, Max | 1.53, 11.46 | 1.13, 11.49 |
| Unknown | 23 | 17 |
| Visit VIS4 | ||
| Mean (SD) | 6.18 (1.73) | 7.29 (1.82) |
| Median (Q1, Q3) | 6.14 (5.05, 7.41) | 7.24 (6.05, 8.54) |
| Min, Max | 0.45, 11.49 | 2.07, 11.47 |
| Unknown | 36 | 18 |
| Visit VIS5 | ||
| Mean (SD) | 6.28 (1.97) | 7.68 (1.94) |
| Median (Q1, Q3) | 6.18 (4.96, 7.71) | 7.75 (6.48, 8.95) |
| Min, Max | 0.87, 11.53 | 2.24, 14.10 |
| Unknown | 40 | 35 |
| Visit VIS6 | ||
| Mean (SD) | 6.69 (1.97) | 8.31 (1.98) |
| Median (Q1, Q3) | 6.64 (5.26, 8.14) | 8.29 (6.92, 9.74) |
| Min, Max | 1.35, 12.95 | 1.93, 14.38 |
| Unknown | 84 | 48 |
| Visit VIS7 | ||
| Mean (SD) | 6.78 (2.09) | 8.78 (2.11) |
| Median (Q1, Q3) | 6.91 (5.46, 8.29) | 8.67 (7.44, 10.26) |
| Min, Max | -1.54, 11.99 | 3.21, 14.36 |
| Unknown | 106 | 78 |
| Visit VIS8 | ||
| Mean (SD) | 7.08 (2.25) | 9.40 (2.26) |
| Median (Q1, Q3) | 7.08 (5.55, 8.68) | 9.35 (7.96, 10.86) |
| Min, Max | 0.97, 13.71 | 2.28, 15.95 |
| Unknown | 123 | 86 |
| Visit VIS9 | ||
| Mean (SD) | 7.39 (2.33) | 10.01 (2.50) |
| Median (Q1, Q3) | 7.47 (5.76, 9.05) | 10.01 (8.19, 11.74) |
| Min, Max | 0.04, 14.61 | 4.22, 18.09 |
| Unknown | 167 | 114 |
| Visit VIS10 | ||
| Mean (SD) | 7.49 (2.58) | 10.59 (2.36) |
| Median (Q1, Q3) | 7.40 (5.73, 9.01) | 10.71 (9.03, 12.25) |
| Min, Max | -0.08, 15.75 | 3.24, 16.40 |
| Unknown | 213 | 170 |
The following figure shows the primary endpoint over the four study visits in the data.
> bcva_data |>
+ group_by(ARMCD) |>
+ ggplot(aes(x = AVISIT, y = BCVA_CHG, fill = factor(ARMCD))) +
+ geom_hline(yintercept = 0, col = "grey", linewidth = 1.2) +
+ geom_boxplot(na.rm = TRUE) +
+ labs(
+ x = "Visit",
+ y = "Change from baseline in BCVA",
+ fill = "Treatment"
+ ) +
+ scale_fill_manual(values = c("darkgoldenrod2", "coral2")) +
+ theme_bw()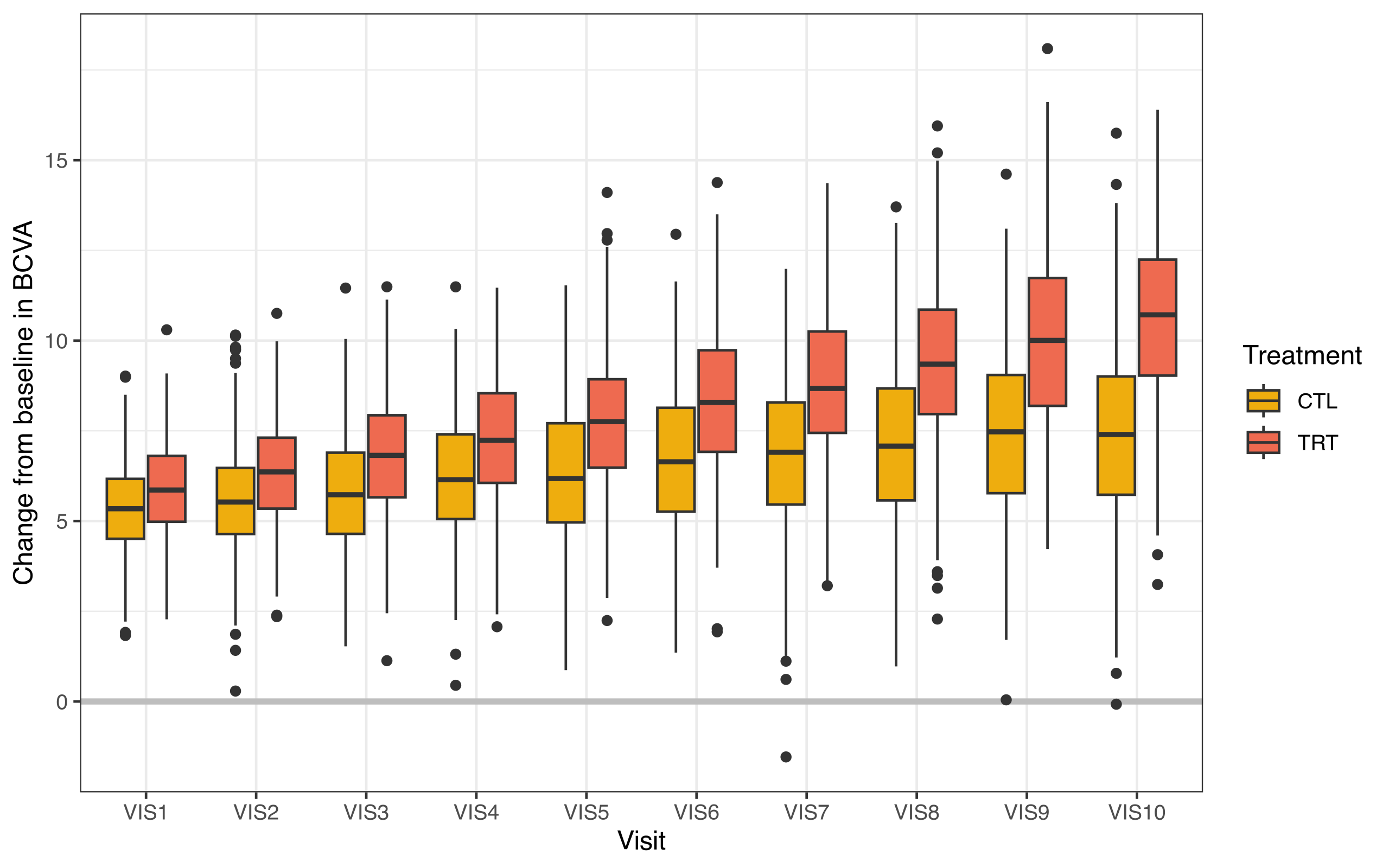
Figure 1. Change from baseline in BCVA over 4 visit time points.
Fitting MMRMs
Bayesian model
The formula for the Bayesian model includes additive effects for baseline, study visit, race, and study-arm-by-visit interaction.
> b_mmrm_formula <- brm_formula(
+ data = bcva_data,
+ intercept = TRUE,
+ baseline = TRUE,
+ group = FALSE,
+ time = TRUE,
+ baseline_time = FALSE,
+ group_time = TRUE,
+ correlation = "unstructured"
+ )
> print(b_mmrm_formula)
#> BCVA_CHG ~ BCVA_BL + ARMCD:AVISIT + AVISIT + RACE + unstr(time = AVISIT, gr = USUBJID)
#> sigma ~ 0 + AVISITWe fit the model using brms.mmrm::brm_model(). The
computation takes several minutes because of the size of the dataset. To
ensure a good basis of comparison with the frequentist model, we put an
extremely diffuse prior on the intercept. The parameters already have
diffuse flexible priors by default.
> b_mmrm_fit <- brm_model(
+ data = filter(bcva_data, !is.na(BCVA_CHG)),
+ formula = b_mmrm_formula,
+ prior = brms::prior(class = "Intercept", prior = "student_t(3, 0, 1000)"),
+ iter = 10000,
+ warmup = 2000,
+ chains = 4,
+ cores = 4,
+ seed = 1,
+ refresh = 0
+ )Here is a posterior summary of model parameters, including fixed effects and pairwise correlation among visits within patients.
> summary(b_mmrm_fit)
#> Family: gaussian
#> Links: mu = identity; sigma = log
#> Formula: BCVA_CHG ~ BCVA_BL + ARMCD:AVISIT + AVISIT + RACE + unstr(time = AVISIT, gr = USUBJID)
#> sigma ~ 0 + AVISIT
#> Data: data[!is.na(data[[attr(data, "brm_outcome")]]), ] (Number of observations: 8605)
#> Draws: 4 chains, each with iter = 10000; warmup = 2000; thin = 1;
#> total post-warmup draws = 32000
#>
#> Correlation Structures:
#> Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
#> cortime(VIS1,VIS2) 0.05 0.03 -0.01 0.11 1.00 63561 23159
#> cortime(VIS1,VIS3) 0.31 0.03 0.25 0.36 1.00 70330 25831
#> cortime(VIS2,VIS3) 0.05 0.03 -0.02 0.11 1.00 67715 22226
#> cortime(VIS1,VIS4) 0.21 0.03 0.15 0.27 1.00 46375 28108
#> cortime(VIS2,VIS4) 0.14 0.03 0.07 0.20 1.00 50232 27277
#> cortime(VIS3,VIS4) -0.01 0.03 -0.07 0.05 1.00 50449 26940
#> cortime(VIS1,VIS5) 0.17 0.03 0.11 0.23 1.00 49366 27023
#> cortime(VIS2,VIS5) 0.12 0.03 0.05 0.18 1.00 53327 28297
#> cortime(VIS3,VIS5) -0.01 0.03 -0.07 0.06 1.00 52752 26884
#> cortime(VIS4,VIS5) 0.38 0.03 0.32 0.43 1.00 49514 26959
#> cortime(VIS1,VIS6) 0.26 0.03 0.20 0.32 1.00 45483 26765
#> cortime(VIS2,VIS6) 0.20 0.03 0.14 0.27 1.00 48236 27168
#> cortime(VIS3,VIS6) 0.04 0.03 -0.02 0.11 1.00 51506 27189
#> cortime(VIS4,VIS6) 0.40 0.03 0.35 0.46 1.00 48730 25696
#> cortime(VIS5,VIS6) 0.39 0.03 0.34 0.45 1.00 55438 25998
#> cortime(VIS1,VIS7) 0.07 0.04 -0.00 0.13 1.00 66961 24586
#> cortime(VIS2,VIS7) 0.09 0.03 0.02 0.15 1.00 66564 23212
#> cortime(VIS3,VIS7) -0.00 0.03 -0.07 0.07 1.00 62299 24284
#> cortime(VIS4,VIS7) 0.15 0.03 0.08 0.22 1.00 70101 23346
#> cortime(VIS5,VIS7) 0.19 0.03 0.13 0.26 1.00 71412 24243
#> cortime(VIS6,VIS7) 0.21 0.04 0.14 0.28 1.00 69307 23697
#> cortime(VIS1,VIS8) 0.05 0.04 -0.02 0.12 1.00 70424 22845
#> cortime(VIS2,VIS8) 0.10 0.04 0.03 0.17 1.00 71230 23497
#> cortime(VIS3,VIS8) -0.03 0.04 -0.10 0.04 1.00 65689 22667
#> cortime(VIS4,VIS8) 0.17 0.03 0.10 0.24 1.00 68079 23681
#> cortime(VIS5,VIS8) 0.17 0.04 0.10 0.24 1.00 73436 24011
#> cortime(VIS6,VIS8) 0.16 0.04 0.09 0.23 1.00 68602 23567
#> cortime(VIS7,VIS8) 0.05 0.04 -0.02 0.13 1.00 68688 23661
#> cortime(VIS1,VIS9) 0.03 0.04 -0.04 0.10 1.00 70389 23613
#> cortime(VIS2,VIS9) -0.01 0.04 -0.08 0.07 1.00 72988 22674
#> cortime(VIS3,VIS9) -0.04 0.04 -0.12 0.03 1.00 73818 23450
#> cortime(VIS4,VIS9) 0.12 0.04 0.04 0.19 1.00 73299 24366
#> cortime(VIS5,VIS9) 0.09 0.04 0.02 0.16 1.00 72264 22069
#> cortime(VIS6,VIS9) 0.17 0.04 0.10 0.24 1.00 74018 24561
#> cortime(VIS7,VIS9) 0.02 0.04 -0.06 0.09 1.00 70521 22326
#> cortime(VIS8,VIS9) 0.06 0.04 -0.02 0.14 1.00 71301 22488
#> cortime(VIS1,VIS10) 0.02 0.04 -0.06 0.10 1.00 62930 25421
#> cortime(VIS2,VIS10) 0.13 0.04 0.05 0.20 1.00 58101 25684
#> cortime(VIS3,VIS10) 0.02 0.04 -0.06 0.10 1.00 60757 24802
#> cortime(VIS4,VIS10) 0.31 0.04 0.24 0.38 1.00 62762 26583
#> cortime(VIS5,VIS10) 0.24 0.04 0.16 0.31 1.00 66606 25076
#> cortime(VIS6,VIS10) 0.30 0.04 0.22 0.37 1.00 67998 23891
#> cortime(VIS7,VIS10) 0.06 0.04 -0.03 0.15 1.00 68944 23170
#> cortime(VIS8,VIS10) 0.09 0.04 0.01 0.18 1.00 71353 23530
#> cortime(VIS9,VIS10) 0.08 0.05 -0.01 0.17 1.00 65710 22799
#>
#> Regression Coefficients:
#> Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
#> Intercept 4.29 0.17 3.96 4.62 1.00 56813
#> BCVA_BL -0.00 0.00 -0.01 0.00 1.00 59119
#> AVISIT2 0.28 0.07 0.14 0.42 1.00 29890
#> AVISIT3 0.46 0.07 0.33 0.59 1.00 44348
#> AVISIT4 0.86 0.08 0.70 1.01 1.00 27610
#> AVISIT5 0.96 0.09 0.79 1.13 1.00 29630
#> AVISIT6 1.33 0.09 1.16 1.50 1.00 28672
#> AVISIT7 1.42 0.11 1.21 1.63 1.00 34514
#> AVISIT8 1.71 0.11 1.49 1.94 1.00 34167
#> AVISIT9 2.00 0.13 1.75 2.25 1.00 35177
#> AVISIT10 2.10 0.14 1.82 2.38 1.00 33084
#> RACEBlack 1.04 0.05 0.93 1.15 1.00 53517
#> RACEWhite 2.01 0.05 1.90 2.11 1.00 54553
#> AVISITVIS1:ARMCDTRT 0.54 0.06 0.41 0.66 1.00 34057
#> AVISITVIS2:ARMCDTRT 0.72 0.08 0.57 0.88 1.00 50542
#> AVISITVIS3:ARMCDTRT 1.01 0.09 0.83 1.19 1.00 48732
#> AVISITVIS4:ARMCDTRT 1.10 0.10 0.91 1.31 1.00 36650
#> AVISITVIS5:ARMCDTRT 1.38 0.12 1.16 1.61 1.00 38946
#> AVISITVIS6:ARMCDTRT 1.63 0.12 1.40 1.86 1.00 36052
#> AVISITVIS7:ARMCDTRT 2.02 0.14 1.74 2.29 1.00 45530
#> AVISITVIS8:ARMCDTRT 2.35 0.15 2.06 2.64 1.00 44496
#> AVISITVIS9:ARMCDTRT 2.66 0.16 2.33 2.98 1.00 44251
#> AVISITVIS10:ARMCDTRT 3.07 0.18 2.71 3.43 1.00 41207
#> sigma_AVISITVIS1 -0.01 0.02 -0.05 0.03 1.00 63843
#> sigma_AVISITVIS2 0.23 0.02 0.18 0.27 1.00 77180
#> sigma_AVISITVIS3 0.36 0.02 0.31 0.40 1.00 68147
#> sigma_AVISITVIS4 0.44 0.02 0.40 0.49 1.00 54719
#> sigma_AVISITVIS5 0.57 0.02 0.52 0.61 1.00 60122
#> sigma_AVISITVIS6 0.58 0.02 0.54 0.63 1.00 54741
#> sigma_AVISITVIS7 0.69 0.02 0.64 0.74 1.00 67848
#> sigma_AVISITVIS8 0.74 0.03 0.69 0.79 1.00 73959
#> sigma_AVISITVIS9 0.80 0.03 0.75 0.85 1.00 73387
#> sigma_AVISITVIS10 0.84 0.03 0.79 0.90 1.00 69664
#> Tail_ESS
#> Intercept 25046
#> BCVA_BL 22844
#> AVISIT2 25900
#> AVISIT3 26347
#> AVISIT4 26145
#> AVISIT5 25959
#> AVISIT6 25061
#> AVISIT7 27504
#> AVISIT8 26821
#> AVISIT9 25947
#> AVISIT10 25296
#> RACEBlack 25805
#> RACEWhite 27113
#> AVISITVIS1:ARMCDTRT 27968
#> AVISITVIS2:ARMCDTRT 25650
#> AVISITVIS3:ARMCDTRT 27016
#> AVISITVIS4:ARMCDTRT 26502
#> AVISITVIS5:ARMCDTRT 25407
#> AVISITVIS6:ARMCDTRT 26418
#> AVISITVIS7:ARMCDTRT 26547
#> AVISITVIS8:ARMCDTRT 26731
#> AVISITVIS9:ARMCDTRT 26034
#> AVISITVIS10:ARMCDTRT 25859
#> sigma_AVISITVIS1 24881
#> sigma_AVISITVIS2 24252
#> sigma_AVISITVIS3 23768
#> sigma_AVISITVIS4 25358
#> sigma_AVISITVIS5 25761
#> sigma_AVISITVIS6 27071
#> sigma_AVISITVIS7 24330
#> sigma_AVISITVIS8 22567
#> sigma_AVISITVIS9 22205
#> sigma_AVISITVIS10 25249
#>
#> Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
#> and Tail_ESS are effective sample size measures, and Rhat is the potential
#> scale reduction factor on split chains (at convergence, Rhat = 1).Frequentist model
The formula for the frequentist model is the same, except for the different syntax for specifying the covariance structure of the MMRM. We fit the model below.
> f_mmrm_fit <- mmrm::mmrm(
+ formula = BCVA_CHG ~ BCVA_BL + ARMCD:AVISIT + AVISIT + RACE +
+ us(AVISIT | USUBJID),
+ data = mutate(
+ bcva_data,
+ AVISIT = factor(as.character(AVISIT), ordered = FALSE)
+ )
+ )The parameter summaries of the frequentist model are below.
> summary(f_mmrm_fit)
#> mmrm fit
#>
#> Formula: BCVA_CHG ~ BCVA_BL + ARMCD:AVISIT + AVISIT + RACE + us(AVISIT |
#> USUBJID)
#> Data:
#> mutate(bcva_data, AVISIT = factor(as.character(AVISIT), ordered = FALSE)) (used
#> 8605 observations from 1000 subjects with maximum 10 timepoints)
#> Covariance: unstructured (55 variance parameters)
#> Method: Satterthwaite
#> Vcov Method: Asymptotic
#> Inference: REML
#>
#> Model selection criteria:
#> AIC BIC logLik deviance
#> 32181.0 32451.0 -16035.5 32071.0
#>
#> Coefficients:
#> Estimate Std. Error df t value Pr(>|t|)
#> (Intercept) 4.288e+00 1.709e-01 1.065e+03 25.085 < 2e-16 ***
#> BCVA_BL -9.935e-04 2.156e-03 9.905e+02 -0.461 0.645
#> AVISITVIS10 2.101e+00 1.400e-01 7.025e+02 15.003 < 2e-16 ***
#> AVISITVIS2 2.810e-01 7.067e-02 9.995e+02 3.976 7.51e-05 ***
#> AVISITVIS3 4.573e-01 6.716e-02 9.747e+02 6.809 1.71e-11 ***
#> AVISITVIS4 8.570e-01 7.636e-02 9.796e+02 11.222 < 2e-16 ***
#> AVISITVIS5 9.638e-01 8.634e-02 9.630e+02 11.163 < 2e-16 ***
#> AVISITVIS6 1.334e+00 8.650e-02 9.451e+02 15.421 < 2e-16 ***
#> AVISITVIS7 1.417e+00 1.071e-01 8.698e+02 13.233 < 2e-16 ***
#> AVISITVIS8 1.711e+00 1.145e-01 8.467e+02 14.944 < 2e-16 ***
#> AVISITVIS9 1.996e+00 1.283e-01 7.784e+02 15.549 < 2e-16 ***
#> RACEBlack 1.038e+00 5.496e-02 1.011e+03 18.891 < 2e-16 ***
#> RACEWhite 2.005e+00 5.198e-02 9.768e+02 38.573 < 2e-16 ***
#> AVISITVIS1:ARMCDTRT 5.391e-01 6.282e-02 9.859e+02 8.582 < 2e-16 ***
#> AVISITVIS10:ARMCDTRT 3.072e+00 1.815e-01 6.620e+02 16.929 < 2e-16 ***
#> AVISITVIS2:ARMCDTRT 7.248e-01 7.984e-02 9.803e+02 9.078 < 2e-16 ***
#> AVISITVIS3:ARMCDTRT 1.012e+00 9.163e-02 9.638e+02 11.039 < 2e-16 ***
#> AVISITVIS4:ARMCDTRT 1.104e+00 1.004e-01 9.653e+02 11.003 < 2e-16 ***
#> AVISITVIS5:ARMCDTRT 1.383e+00 1.147e-01 9.505e+02 12.065 < 2e-16 ***
#> AVISITVIS6:ARMCDTRT 1.630e+00 1.189e-01 9.157e+02 13.715 < 2e-16 ***
#> AVISITVIS7:ARMCDTRT 2.016e+00 1.382e-01 8.262e+02 14.592 < 2e-16 ***
#> AVISITVIS8:ARMCDTRT 2.347e+00 1.474e-01 8.041e+02 15.924 < 2e-16 ***
#> AVISITVIS9:ARMCDTRT 2.658e+00 1.644e-01 7.277e+02 16.172 < 2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Covariance estimate:
#> VIS1 VIS10 VIS2 VIS3 VIS4 VIS5 VIS6 VIS7 VIS8
#> VIS1 0.9713 0.0587 0.0630 0.4371 0.3315 0.3056 0.4688 0.1325 0.1020
#> VIS10 0.0587 5.3519 0.3761 0.0719 1.1478 0.9997 1.2558 0.3021 0.4658
#> VIS2 0.0630 0.3761 1.5618 0.0871 0.2684 0.2635 0.4636 0.2180 0.2776
#> VIS3 0.4371 0.0719 0.0871 2.0221 -0.0216 -0.0189 0.1102 -0.0048 -0.0993
#> VIS4 0.3315 1.1478 0.2684 -0.0216 2.4113 1.0475 1.1409 0.4625 0.5659
#> VIS5 0.3056 0.9997 0.2635 -0.0189 1.0475 3.0916 1.2593 0.6911 0.6308
#> VIS6 0.4688 1.2558 0.4636 0.1102 1.1409 1.2593 3.1853 0.7540 0.6094
#> VIS7 0.1325 0.3021 0.2180 -0.0048 0.4625 0.6911 0.7540 3.9272 0.2306
#> VIS8 0.1020 0.4658 0.2776 -0.0993 0.5659 0.6308 0.6094 0.2306 4.3272
#> VIS9 0.0611 0.4141 -0.0153 -0.1321 0.4085 0.3594 0.6823 0.0723 0.2683
#> VIS9
#> VIS1 0.0611
#> VIS10 0.4141
#> VIS2 -0.0153
#> VIS3 -0.1321
#> VIS4 0.4085
#> VIS5 0.3594
#> VIS6 0.6823
#> VIS7 0.0723
#> VIS8 0.2683
#> VIS9 4.8635Comparison
This section compares the Bayesian posterior parameter estimates from
brms.mmrm to the frequentist parameter estimates of the
mmrm package.
Extract estimates from Bayesian model
We extract and standardize the Bayesian estimates.
> b_mmrm_draws <- b_mmrm_fit |>
+ as_draws_df()
> visit_levels <- sort(unique(as.character(bcva_data$AVISIT)))
> for (level in visit_levels) {
+ name <- paste0("b_sigma_AVISIT", level)
+ b_mmrm_draws[[name]] <- exp(b_mmrm_draws[[name]])
+ }
> b_mmrm_summary <- b_mmrm_draws |>
+ summarize_draws() |>
+ select(variable, mean, sd) |>
+ filter(!(variable %in% c("Intercept", "lprior", "lp__"))) |>
+ rename(bayes_estimate = mean, bayes_se = sd) |>
+ mutate(
+ variable = variable |>
+ tolower() |>
+ gsub(pattern = "b_", replacement = "") |>
+ gsub(pattern = "b_sigma_AVISIT", replacement = "sigma_") |>
+ gsub(pattern = "cortime", replacement = "correlation") |>
+ gsub(pattern = "__", replacement = "_") |>
+ gsub(pattern = "avisitvis", replacement = "avisit")
+ )Extract estimates from frequentist model
We extract and standardize the frequentist estimates.
> f_mmrm_fixed <- summary(f_mmrm_fit)$coefficients |>
+ as_tibble(rownames = "variable") |>
+ mutate(variable = tolower(variable)) |>
+ mutate(variable = gsub("(", "", variable, fixed = TRUE)) |>
+ mutate(variable = gsub(")", "", variable, fixed = TRUE)) |>
+ mutate(variable = gsub("avisitvis", "avisit", variable)) |>
+ rename(freq_estimate = Estimate, freq_se = `Std. Error`) |>
+ select(variable, freq_estimate, freq_se)> f_mmrm_variance <- tibble(
+ variable = paste0("sigma_AVISIT", visit_levels) |>
+ tolower() |>
+ gsub(pattern = "avisitvis", replacement = "avisit"),
+ freq_estimate = sqrt(diag(f_mmrm_fit$cov))
+ )> f_diagonal_factor <- diag(1 / sqrt(diag(f_mmrm_fit$cov)))
> f_corr_matrix <- f_diagonal_factor %*% f_mmrm_fit$cov %*% f_diagonal_factor
> colnames(f_corr_matrix) <- visit_levels> f_mmrm_correlation <- f_corr_matrix |>
+ as.data.frame() |>
+ as_tibble() |>
+ mutate(x1 = visit_levels) |>
+ pivot_longer(
+ cols = -any_of("x1"),
+ names_to = "x2",
+ values_to = "freq_estimate"
+ ) |>
+ filter(
+ as.numeric(gsub("[^0-9]", "", x1)) < as.numeric(gsub("[^0-9]", "", x2))
+ ) |>
+ mutate(variable = sprintf("correlation_%s_%s", x1, x2)) |>
+ select(variable, freq_estimate)Summary
The first table below summarizes the parameter estimates from each model and the differences between estimates (Bayesian minus frequentist). The second table shows the standard errors of these estimates and differences between standard errors. In each table, the “Relative” column shows the relative difference (the difference divided by the frequentist quantity).
Because of the different statistical paradigms and estimation
procedures, especially regarding the covariance parameters, it would not
be realistic to expect the Bayesian and frequentist approaches to yield
virtually identical results. Nevertheless, the absolute and relative
differences in the table below show strong agreement between
brms.mmrm and mmrm.
> b_f_comparison <- full_join(
+ x = b_mmrm_summary,
+ y = f_mmrm_summary,
+ by = "variable"
+ ) |>
+ mutate(
+ diff_estimate = bayes_estimate - freq_estimate,
+ diff_relative_estimate = diff_estimate / freq_estimate,
+ diff_se = bayes_se - freq_se,
+ diff_relative_se = diff_se / freq_se
+ ) |>
+ select(variable, ends_with("estimate"), ends_with("se"))> table_estimates <- b_f_comparison |>
+ select(variable, ends_with("estimate"))
> gt(table_estimates) |>
+ fmt_number(decimals = 4) |>
+ tab_caption(
+ caption = md(
+ paste(
+ "Table 4. Comparison of parameter estimates between",
+ "Bayesian and frequentist MMRMs."
+ )
+ )
+ ) |>
+ cols_label(
+ variable = "Variable",
+ bayes_estimate = "Bayesian",
+ freq_estimate = "Frequentist",
+ diff_estimate = "Difference",
+ diff_relative_estimate = "Relative"
+ )| Variable | Bayesian | Frequentist | Difference | Relative |
|---|---|---|---|---|
| intercept | 4.2889 | 4.2881 | 0.0009 | 0.0002 |
| bcva_bl | −0.0010 | −0.0010 | 0.0000 | 0.0143 |
| avisit2 | 0.2806 | 0.2810 | −0.0004 | −0.0014 |
| avisit3 | 0.4577 | 0.4573 | 0.0005 | 0.0010 |
| avisit4 | 0.8564 | 0.8570 | −0.0005 | −0.0006 |
| avisit5 | 0.9631 | 0.9638 | −0.0007 | −0.0007 |
| avisit6 | 1.3333 | 1.3339 | −0.0006 | −0.0005 |
| avisit7 | 1.4161 | 1.4167 | −0.0006 | −0.0005 |
| avisit8 | 1.7106 | 1.7107 | −0.0001 | −0.0001 |
| avisit9 | 1.9955 | 1.9956 | −0.0001 | 0.0000 |
| avisit10 | 2.0997 | 2.1005 | −0.0008 | −0.0004 |
| raceblack | 1.0385 | 1.0382 | 0.0002 | 0.0002 |
| racewhite | 2.0054 | 2.0051 | 0.0003 | 0.0002 |
| avisit1:armcdtrt | 0.5391 | 0.5391 | 0.0000 | −0.0001 |
| avisit2:armcdtrt | 0.7249 | 0.7248 | 0.0001 | 0.0001 |
| avisit3:armcdtrt | 1.0110 | 1.0115 | −0.0005 | −0.0005 |
| avisit4:armcdtrt | 1.1049 | 1.1042 | 0.0007 | 0.0007 |
| avisit5:armcdtrt | 1.3843 | 1.3834 | 0.0009 | 0.0007 |
| avisit6:armcdtrt | 1.6304 | 1.6301 | 0.0003 | 0.0002 |
| avisit7:armcdtrt | 2.0168 | 2.0160 | 0.0009 | 0.0004 |
| avisit8:armcdtrt | 2.3471 | 2.3469 | 0.0002 | 0.0001 |
| avisit9:armcdtrt | 2.6592 | 2.6585 | 0.0007 | 0.0003 |
| avisit10:armcdtrt | 3.0742 | 3.0723 | 0.0019 | 0.0006 |
| sigma_avisit1 | 0.9893 | 0.9855 | 0.0037 | 0.0038 |
| sigma_avisit2 | 1.2557 | 1.2497 | 0.0060 | 0.0048 |
| sigma_avisit3 | 1.4289 | 1.4220 | 0.0069 | 0.0048 |
| sigma_avisit4 | 1.5568 | 1.5528 | 0.0040 | 0.0026 |
| sigma_avisit5 | 1.7633 | 1.7583 | 0.0050 | 0.0028 |
| sigma_avisit6 | 1.7888 | 1.7847 | 0.0041 | 0.0023 |
| sigma_avisit7 | 1.9931 | 1.9817 | 0.0113 | 0.0057 |
| sigma_avisit8 | 2.0922 | 2.0802 | 0.0120 | 0.0058 |
| sigma_avisit9 | 2.2208 | 2.2053 | 0.0155 | 0.0070 |
| sigma_avisit10 | 2.3279 | 2.3134 | 0.0145 | 0.0063 |
| correlation_vis1_vis2 | 0.0489 | 0.0512 | −0.0023 | −0.0441 |
| correlation_vis1_vis3 | 0.3084 | 0.3119 | −0.0036 | −0.0114 |
| correlation_vis2_vis3 | 0.0482 | 0.0490 | −0.0008 | −0.0164 |
| correlation_vis1_vis4 | 0.2126 | 0.2166 | −0.0040 | −0.0184 |
| correlation_vis2_vis4 | 0.1351 | 0.1383 | −0.0033 | −0.0237 |
| correlation_vis3_vis4 | −0.0106 | −0.0098 | −0.0008 | 0.0869 |
| correlation_vis1_vis5 | 0.1722 | 0.1764 | −0.0041 | −0.0234 |
| correlation_vis2_vis5 | 0.1167 | 0.1199 | −0.0032 | −0.0265 |
| correlation_vis3_vis5 | −0.0082 | −0.0076 | −0.0006 | 0.0849 |
| correlation_vis4_vis5 | 0.3770 | 0.3836 | −0.0066 | −0.0173 |
| correlation_vis1_vis6 | 0.2617 | 0.2665 | −0.0048 | −0.0181 |
| correlation_vis2_vis6 | 0.2038 | 0.2079 | −0.0040 | −0.0194 |
| correlation_vis3_vis6 | 0.0422 | 0.0434 | −0.0012 | −0.0279 |
| correlation_vis4_vis6 | 0.4044 | 0.4117 | −0.0073 | −0.0177 |
| correlation_vis5_vis6 | 0.3941 | 0.4013 | −0.0072 | −0.0179 |
| correlation_vis1_vis7 | 0.0654 | 0.0679 | −0.0024 | −0.0360 |
| correlation_vis2_vis7 | 0.0857 | 0.0880 | −0.0023 | −0.0266 |
| correlation_vis3_vis7 | −0.0019 | −0.0017 | −0.0002 | 0.1039 |
| correlation_vis4_vis7 | 0.1464 | 0.1503 | −0.0040 | −0.0263 |
| correlation_vis5_vis7 | 0.1941 | 0.1983 | −0.0042 | −0.0214 |
| correlation_vis6_vis7 | 0.2083 | 0.2132 | −0.0048 | −0.0227 |
| correlation_vis1_vis8 | 0.0478 | 0.0497 | −0.0019 | −0.0382 |
| correlation_vis2_vis8 | 0.1044 | 0.1068 | −0.0024 | −0.0225 |
| correlation_vis3_vis8 | −0.0332 | −0.0336 | 0.0004 | −0.0112 |
| correlation_vis4_vis8 | 0.1712 | 0.1752 | −0.0040 | −0.0229 |
| correlation_vis5_vis8 | 0.1683 | 0.1725 | −0.0041 | −0.0240 |
| correlation_vis6_vis8 | 0.1597 | 0.1641 | −0.0045 | −0.0273 |
| correlation_vis7_vis8 | 0.0538 | 0.0559 | −0.0022 | −0.0392 |
| correlation_vis1_vis9 | 0.0269 | 0.0281 | −0.0012 | −0.0432 |
| correlation_vis2_vis9 | −0.0065 | −0.0056 | −0.0010 | 0.1708 |
| correlation_vis3_vis9 | −0.0416 | −0.0421 | 0.0005 | −0.0124 |
| correlation_vis4_vis9 | 0.1160 | 0.1193 | −0.0033 | −0.0273 |
| correlation_vis5_vis9 | 0.0898 | 0.0927 | −0.0029 | −0.0313 |
| correlation_vis6_vis9 | 0.1692 | 0.1733 | −0.0041 | −0.0238 |
| correlation_vis7_vis9 | 0.0153 | 0.0165 | −0.0013 | −0.0761 |
| correlation_vis8_vis9 | 0.0569 | 0.0585 | −0.0016 | −0.0267 |
| correlation_vis1_vis10 | 0.0229 | 0.0257 | −0.0029 | −0.1112 |
| correlation_vis2_vis10 | 0.1266 | 0.1301 | −0.0035 | −0.0267 |
| correlation_vis3_vis10 | 0.0217 | 0.0219 | −0.0002 | −0.0070 |
| correlation_vis4_vis10 | 0.3115 | 0.3195 | −0.0080 | −0.0251 |
| correlation_vis5_vis10 | 0.2385 | 0.2458 | −0.0073 | −0.0298 |
| correlation_vis6_vis10 | 0.2959 | 0.3041 | −0.0082 | −0.0271 |
| correlation_vis7_vis10 | 0.0631 | 0.0659 | −0.0028 | −0.0422 |
| correlation_vis8_vis10 | 0.0932 | 0.0968 | −0.0037 | −0.0377 |
| correlation_vis9_vis10 | 0.0781 | 0.0812 | −0.0031 | −0.0383 |
> table_se <- b_f_comparison |>
+ select(variable, ends_with("se")) |>
+ filter(!is.na(freq_se))
> gt(table_se) |>
+ fmt_number(decimals = 4) |>
+ tab_caption(
+ caption = md(
+ paste(
+ "Table 5. Comparison of parameter standard errors between",
+ "Bayesian and frequentist MMRMs."
+ )
+ )
+ ) |>
+ cols_label(
+ variable = "Variable",
+ bayes_se = "Bayesian",
+ freq_se = "Frequentist",
+ diff_se = "Difference",
+ diff_relative_se = "Relative"
+ )| Variable | Bayesian | Frequentist | Difference | Relative |
|---|---|---|---|---|
| intercept | 0.1695 | 0.1709 | −0.0015 | −0.0086 |
| bcva_bl | 0.0021 | 0.0022 | 0.0000 | −0.0100 |
| avisit2 | 0.0709 | 0.0707 | 0.0003 | 0.0038 |
| avisit3 | 0.0675 | 0.0672 | 0.0003 | 0.0052 |
| avisit4 | 0.0771 | 0.0764 | 0.0007 | 0.0094 |
| avisit5 | 0.0868 | 0.0863 | 0.0005 | 0.0055 |
| avisit6 | 0.0869 | 0.0865 | 0.0004 | 0.0042 |
| avisit7 | 0.1081 | 0.1071 | 0.0011 | 0.0102 |
| avisit8 | 0.1147 | 0.1145 | 0.0002 | 0.0017 |
| avisit9 | 0.1276 | 0.1283 | −0.0007 | −0.0057 |
| avisit10 | 0.1418 | 0.1400 | 0.0018 | 0.0130 |
| raceblack | 0.0548 | 0.0550 | −0.0001 | −0.0024 |
| racewhite | 0.0518 | 0.0520 | −0.0001 | −0.0029 |
| avisit1:armcdtrt | 0.0632 | 0.0628 | 0.0003 | 0.0054 |
| avisit2:armcdtrt | 0.0806 | 0.0798 | 0.0007 | 0.0093 |
| avisit3:armcdtrt | 0.0925 | 0.0916 | 0.0008 | 0.0092 |
| avisit4:armcdtrt | 0.1017 | 0.1004 | 0.0014 | 0.0136 |
| avisit5:armcdtrt | 0.1157 | 0.1147 | 0.0010 | 0.0088 |
| avisit6:armcdtrt | 0.1189 | 0.1189 | 0.0000 | 0.0003 |
| avisit7:armcdtrt | 0.1390 | 0.1382 | 0.0008 | 0.0060 |
| avisit8:armcdtrt | 0.1484 | 0.1474 | 0.0010 | 0.0066 |
| avisit9:armcdtrt | 0.1643 | 0.1644 | −0.0001 | −0.0004 |
| avisit10:armcdtrt | 0.1837 | 0.1815 | 0.0022 | 0.0122 |