
Plotting dose-toxicity and dose-biomarker model fits
Source:R/Samples-methods.R
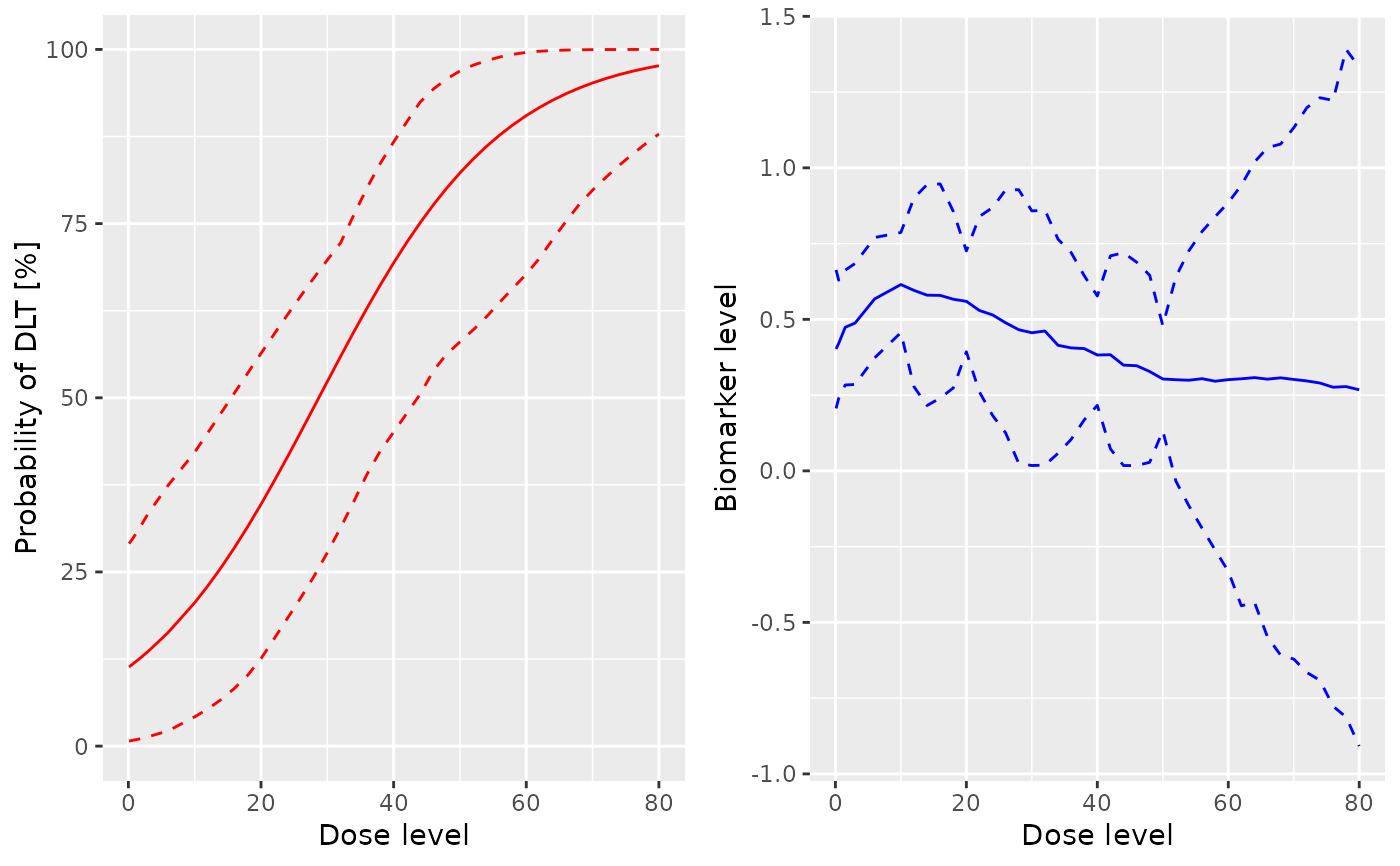
plot-Samples-DualEndpoint-method.RdWhen we have the dual endpoint model, also the dose-biomarker fit is shown in the plot
Usage
# S4 method for class 'Samples,DualEndpoint'
plot(x, y, data, extrapolate = TRUE, showLegend = FALSE, ...)Arguments
- x
the
Samplesobject- y
the
DualEndpointobject- data
the
DataDualobject- extrapolate
should the biomarker fit be extrapolated to the whole dose grid? (default)
- showLegend
should the legend be shown? (not default)
- ...
additional arguments for the parent method
plot,Samples,GeneralModel-method
Value
This returns the ggplot
object with the dose-toxicity and dose-biomarker model fits
Examples
# nolint start
# Create some data
data <- DataDual(
x = c(0.1, 0.5, 1.5, 3, 6, 10, 10, 10, 20, 20, 20, 40, 40, 40, 50, 50, 50),
y = c(0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 1, 0, 0, 1, 0, 1, 1),
w = c(
0.31,
0.42,
0.59,
0.45,
0.6,
0.7,
0.55,
0.6,
0.52,
0.54,
0.56,
0.43,
0.41,
0.39,
0.34,
0.38,
0.21
),
doseGrid = c(0.1, 0.5, 1.5, 3, 6, seq(from = 10, to = 80, by = 2))
)
#> Used default patient IDs!
#> Used best guess cohort indices!
# Initialize the Dual-Endpoint model (in this case RW1)
model <- DualEndpointRW(
mean = c(0, 1),
cov = matrix(c(1, 0, 0, 1), nrow = 2),
sigma2betaW = 0.01,
sigma2W = c(a = 0.1, b = 0.1),
rho = c(a = 1, b = 1),
rw1 = TRUE
)
# Set-up some MCMC parameters and generate samples from the posterior
options <- McmcOptions(burnin = 100, step = 2, samples = 500)
set.seed(94)
samples <- mcmc(data, model, options)
# Plot the posterior mean (and empirical 2.5 and 97.5 percentile)
# for the prob(DLT) by doses and the Biomarker by doses
#grid.arrange(plot(x = samples, y = model, data = data))
plot(x = samples, y = model, data = data)
 # nolint end
# nolint end