
Rolling CRM Example
Jiawen Zhu, Daniel Sabanes Bove
02 January 2019
Source:vignettes/rolling-crm.Rmd
rolling-crm.RmdExample 1: Recommend a dose for the next cohort
Setting up the data
library(crmPack)
data <- DataDA(
x = c(0.1, 0.5, 1.5, 3, 6, 10, 10, 10),
y = c(0, 0, 1, 1, 0, 0, 1, 0),
ID = as.integer(1:8),
cohort = as.integer(c(1, 2, 3, 4, 5, 6, 6, 6)),
doseGrid =
c(
0.1, 0.5, 1.5, 3, 6,
seq(from = 10, to = 80, by = 2)
),
u = c(42, 30, 15, 5, 20, 25, 30, 60),
t0 = rep(0, 8),
Tmax = 60
)
emptydata <- DataDA(
doseGrid = c(
0.1, 0.5, 1, 1.5, 3, 6,
seq(from = 10, to = 80, by = 2)
),
Tmax = 60
)Structure of the model class
npiece_ <- 10
Tmax_ <- 60
lambda_prior <- function(k) {
npiece_ / (Tmax_ * (npiece_ - k + 0.5))
}
model <- DALogisticLogNormal(
mean = c(-0.85, 1),
cov = matrix(c(1, -0.5, -0.5, 1), nrow = 2),
ref_dose = 56,
npiece = npiece_,
l = as.numeric(t(apply(as.matrix(c(1:npiece_), 1, npiece_), 2, lambda_prior))),
c_par = 2
)Obtain the posterior
options <- McmcOptions(
burnin = 10,
step = 2,
samples = 1e2
)
set.seed(94)
samples <- mcmc(data, model, options)Use ggmcmc to diagnose
library(ggmcmc)
alpha0samples <- get(samples, "alpha0")
print(ggs_traceplot(alpha0samples))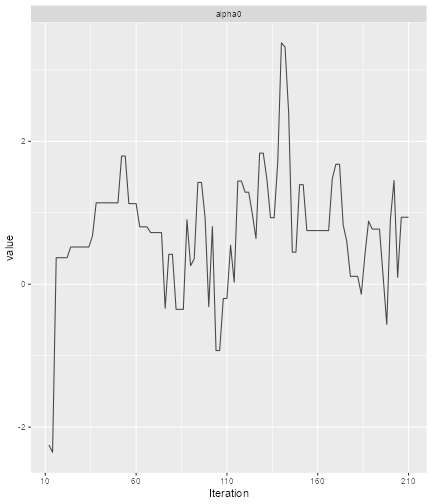
plot of chunk Diagnose-1
print(ggs_autocorrelation(alpha0samples))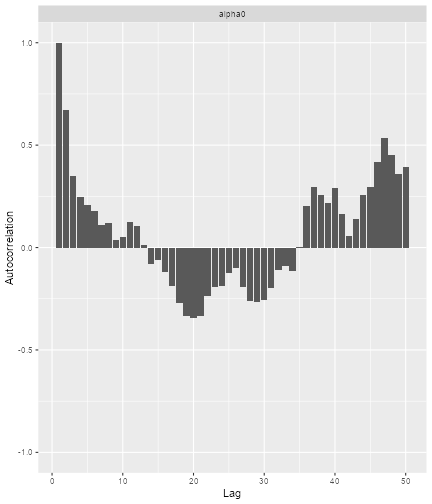
plot of chunk Diagnose-2
Plot the model fit
plot(samples, model, data, hazard = TRUE)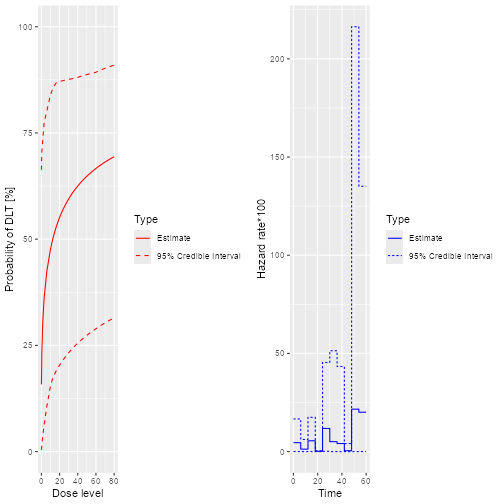
plot of chunk Fit-1
plot(samples, model, data, hazard = FALSE)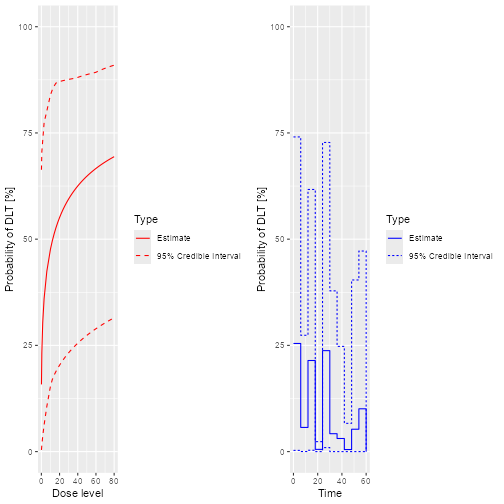
plot of chunk Fit-2
prior mean curve
emptydata <- DataDA(doseGrid = c(
0.1, 0.5, 1.5, 3, 6,
seq(from = 10, to = 80, by = 2)
), Tmax = 60)
Priorsamples <- mcmc(emptydata, model, options)
plot(Priorsamples, model, emptydata, hazard = FALSE)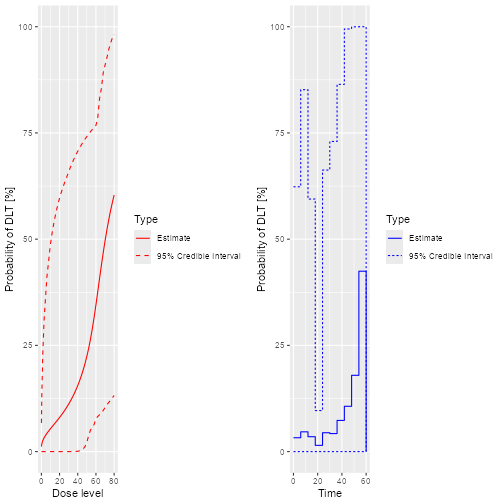
plot of chunk Prior
Escalation rules
Need to fill in (use the same rule in the section 8 of “using the package crmPack: introductory examples”)
myIncrements <- IncrementsRelative(
intervals = c(0, 20),
increments = c(1, 0.33)
)
nextMaxDose <- maxDose(myIncrements, data = data)
myNextBest <- NextBestNCRM(
target = c(0.2, 0.35),
overdose = c(0.35, 1),
max_overdose_prob = 0.25
)
mySize1 <- CohortSizeRange(intervals = c(0, 30), cohort_size = c(1, 3))
mySize2 <- CohortSizeDLT(intervals = c(0, 1), cohort_size = c(1, 3))
mySize <- maxSize(mySize1, mySize2)
myStopping1 <- StoppingTargetProb(target = c(0.2, 0.35), prob = 0.5)
myStopping2 <- StoppingMinPatients(nPatients = 50)
myStopping <- (myStopping1 | myStopping2)Recommended dose for the next cohort
doseRecommendation <- nextBest(myNextBest,
doselimit = nextMaxDose,
samples = samples,
model = model,
data = data
)
doseRecommendation$plot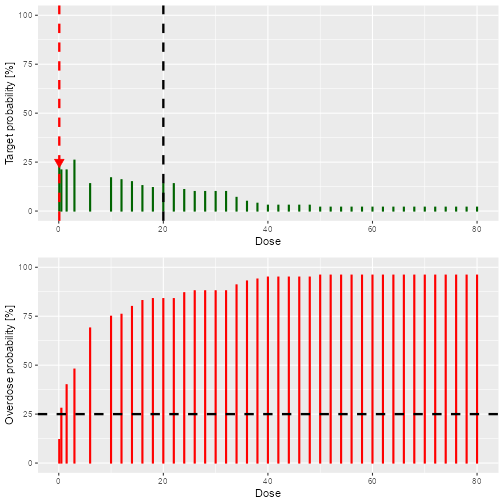
plot of chunk Recommend
doseRecommendation$value
#> [1] 0.1Example 2: Run a simulation to evaluate operating characteristics
Set up safety window and DADesign to be completed
mysafetywindow <- SafetyWindowConst(c(6, 2), 7, 7)
design <- DADesign(
model = model,
increments = myIncrements,
nextBest = myNextBest,
stopping = myStopping,
cohort_size = mySize,
data = emptydata,
safetyWindow = mysafetywindow,
startingDose = 3
)Set up true curves
myTruth <- probFunction(model, alpha0 = 2, alpha1 = 3)
curve(myTruth(x), from = 0, to = 100, ylim = c(0, 1))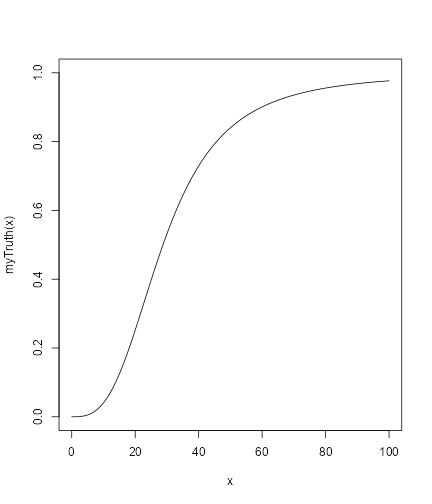
plot of chunk Truth
Perform the simulations
mySims <- simulate(design,
args = NULL,
truthTox = myTruth,
truthSurv = exp_cond.cdf,
trueTmax = 80,
nsim = 2,
seed = 819,
mcmcOptions = options,
firstSeparate = TRUE,
deescalate = FALSE,
parallel = FALSE
)Interpret the simulation results
Use a similar way as section 9.2 in the “using the package crmPack: introductory examples” document
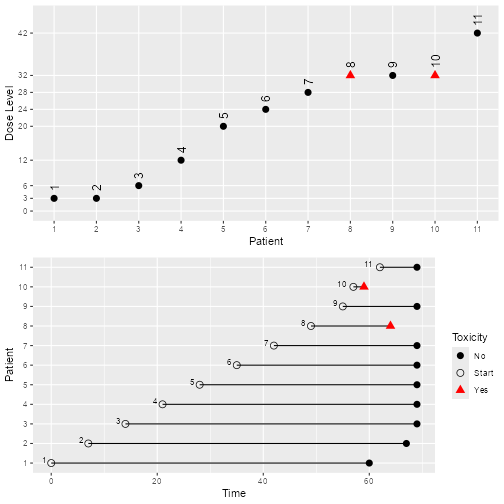
plot of chunk Interpret
mySims@stop_reasons[[2]]
#> [[1]]
#> [1] "Probability for target toxicity is 68 % for dose 34 and thus above the required 50 %"
#>
#> [[2]]
#> [1] "Number of patients is 17 and thus below the prespecified minimum number 50"
# nolint end