
Plot the fitted dose-efficacy curve using a model from ModelEff class with samples
Source: R/Samples-methods.R
plot-Samples-ModelEff-method.RdPlot the fitted dose-efficacy curve using a model from ModelEff class
with samples
Usage
# S4 method for class 'Samples,ModelEff'
plot(
x,
y,
data,
...,
xlab = "Dose level",
ylab = "Expected Efficacy",
showLegend = TRUE
)Value
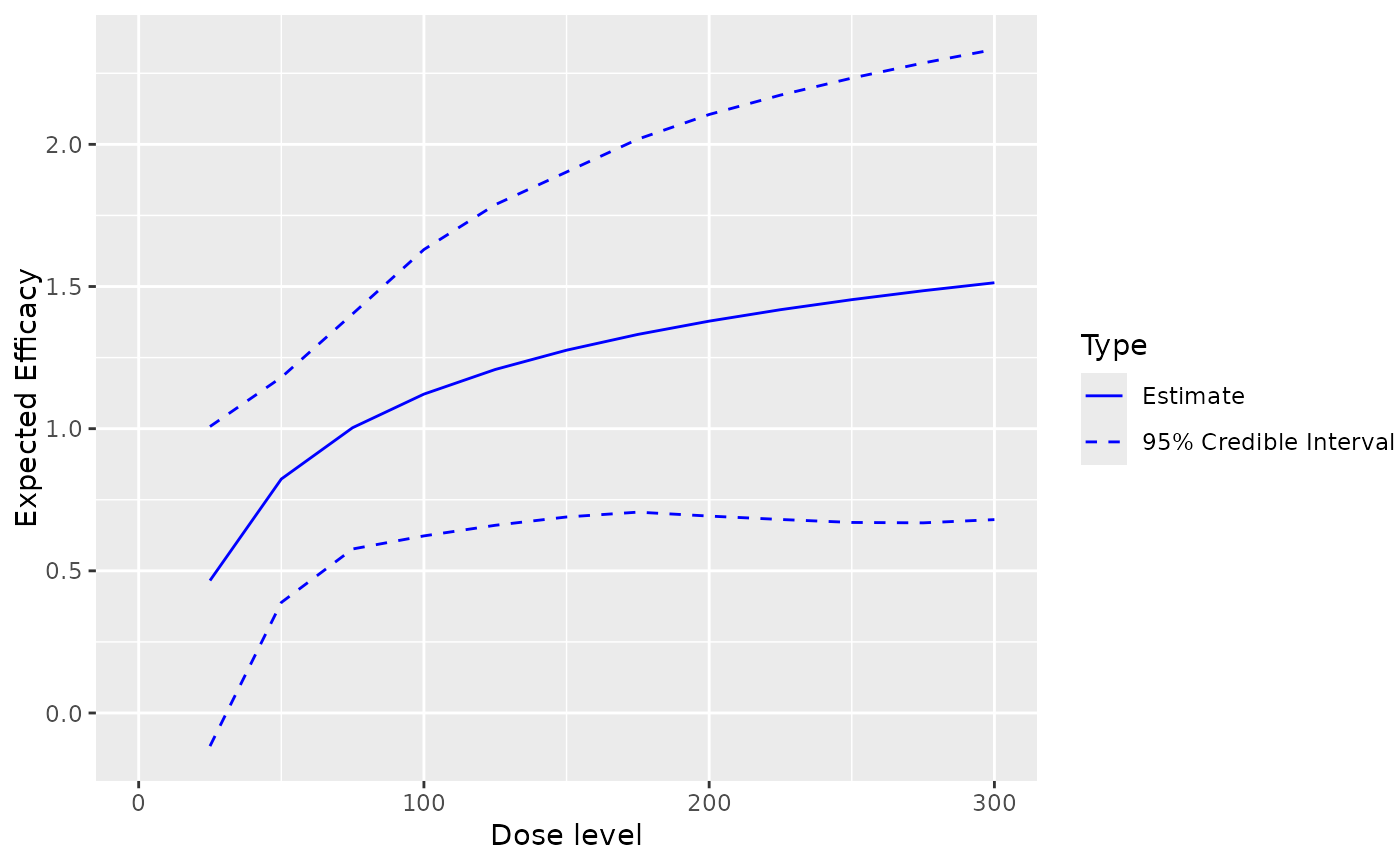
This returns the ggplot
object for the dose-efficacy model fit
Examples
# nolint start
## we need a data object with doses >= 1:
data <- DataDual(
x = c(25, 50, 25, 50, 75, 300, 250, 150),
y = c(0, 0, 0, 0, 0, 1, 1, 0),
w = c(0.31, 0.42, 0.59, 0.45, 0.6, 0.7, 0.6, 0.52),
doseGrid = seq(25, 300, 25),
placebo = FALSE
)
#> Used default patient IDs!
#> Used best guess cohort indices!
##plot the dose-efficacy curve with samples using the model from 'ModelEff'
##class e.g. 'Effloglog' class model
##define the model (see Effloglog example)
Effmodel <- Effloglog(
eff = c(1.223, 2.513),
eff_dose = c(25, 300),
nu = c(a = 1, b = 0.025),
data = data
)
## define the samples obtained using the 'Effloglog' model (see details in 'Samples' example)
##options for MCMC
options <- McmcOptions(burnin = 100, step = 2, samples = 200)
## samples must be of 'Samples' class
samples <- mcmc(data = data, model = Effmodel, options = options)
## plot the fitted dose-efficacy curve including the 95% credibility interval of the samples
## 'x' should be of 'Samples' class and 'y' of 'ModelEff' class
plot(x = samples, y = Effmodel, data = data)
 # nolint end
# nolint end