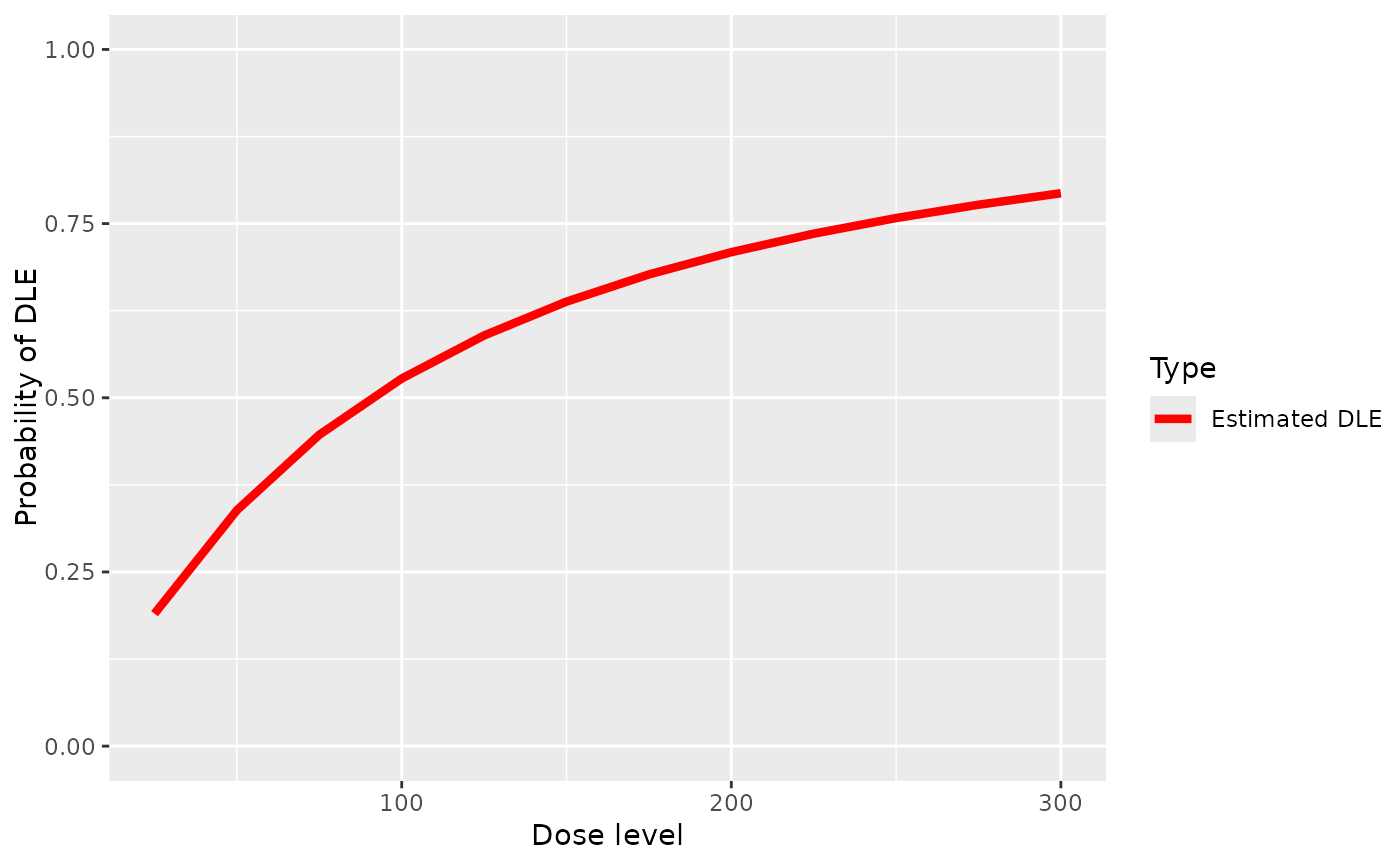
Plot of the fitted dose-tox based with a given pseudo DLE model and data without samples
Source:R/Samples-methods.R
plot-Data-ModelTox-method.RdPlot of the fitted dose-tox based with a given pseudo DLE model and data without samples
Usage
# S4 method for class 'Data,ModelTox'
plot(
x,
y,
xlab = "Dose level",
ylab = "Probability of DLE",
showLegend = TRUE,
...
)Value
This returns the ggplot
object for the dose-DLE model plot
Examples
## plot the dose-DLE curve given a pseudo DLE model using data without samples
## data must be of 'Data' class
## define the data
data <- Data(
x = c(25, 50, 50, 75, 100, 100, 225, 300),
y = c(0, 0, 0, 0, 1, 1, 1, 1),
ID = 1L:8L,
cohort = as.integer(c(1, 2, 2, 3, 4, 4, 5, 6)),
doseGrid = seq(25, 300, 25)
)
## model must be from 'ModelTox' class e.g 'LogisticIndepBeta' class model
## define the model (see LogisticIndepBeta example)
model <- LogisticIndepBeta(
binDLE = c(1.05, 1.8),
DLEweights = c(3, 3),
DLEdose = c(25, 300),
data = data
)
## plot the dose-DLE curve
## 'x' is the data and 'y' is the model in plot
plot(x = data, y = model)