
Plot Model-Based Design Simulation Summary
Source:R/Simulations-methods.R
plot-SimulationsSummary-missing-method.RdGraphical display of the simulation summary.
This plot method can be applied to SimulationsSummary objects in order
to summarize them graphically. Possible type of plots at the moment are
those listed in plot,GeneralSimulationsSummary,missing-method plus:
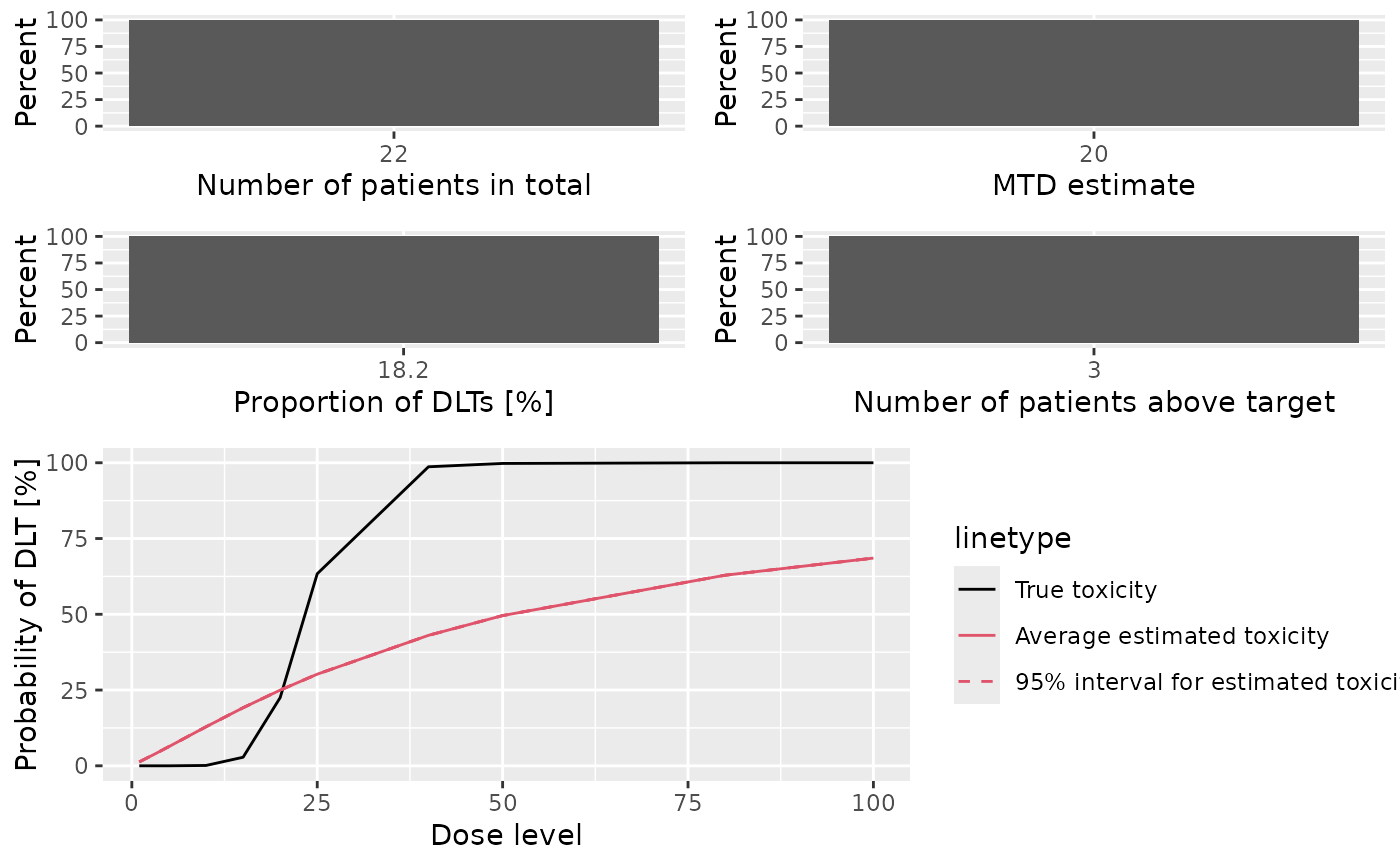
- meanFit
Plot showing the average fitted dose-toxicity curve across the trials, together with 95% credible intervals, and comparison with the assumed truth (as specified by the
truthargument tosummary,Simulations-method)
You can specify any subset of these in the type argument.
Examples
# nolint start
# Define the dose-grid
emptydata <- Data(doseGrid = c(1, 3, 5, 10, 15, 20, 25, 40, 50, 80, 100))
# Initialize the CRM model
model <- LogisticLogNormal(
mean = c(-0.85, 1),
cov = matrix(c(1, -0.5, -0.5, 1), nrow = 2),
ref_dose = 56
)
# Choose the rule for selecting the next dose
myNextBest <- NextBestNCRM(
target = c(0.2, 0.35),
overdose = c(0.35, 1),
max_overdose_prob = 0.25
)
# Choose the rule for the cohort-size
mySize1 <- CohortSizeRange(
intervals = c(0, 30),
cohort_size = c(1, 3)
)
mySize2 <- CohortSizeDLT(
intervals = c(0, 1),
cohort_size = c(1, 3)
)
mySize <- maxSize(mySize1, mySize2)
# Choose the rule for stopping
myStopping1 <- StoppingMinCohorts(nCohorts = 3)
myStopping2 <- StoppingTargetProb(
target = c(0.2, 0.35),
prob = 0.5
)
myStopping3 <- StoppingMinPatients(nPatients = 20)
myStopping <- (myStopping1 & myStopping2) | myStopping3 | StoppingMissingDose()
# Choose the rule for dose increments
myIncrements <- IncrementsRelative(
intervals = c(0, 20),
increments = c(1, 0.33)
)
# Initialize the design
design <- Design(
model = model,
nextBest = myNextBest,
stopping = myStopping,
increments = myIncrements,
cohort_size = mySize,
data = emptydata,
startingDose = 3
)
## define the true function
myTruth <- probFunction(model, alpha0 = 7, alpha1 = 8)
# Run the simulation on the desired design
# We only generate 1 trial outcomes here for illustration, for the actual study
# this should be increased of course
options <- McmcOptions(
burnin = 5,
step = 1,
samples = 10
)
time <- system.time(
mySims <- simulate(
design,
args = NULL,
truth = myTruth,
nsim = 1,
seed = 819,
mcmcOptions = options,
parallel = FALSE
)
)[3]
# Plot the Summary of the Simulations
plot(summary(mySims, truth = myTruth))
 # nolint end
# nolint end